Our Team
Jean Philippe Combier
PI, CNRS
jean-philippe.combier@univ-tlse3.fr

Serge Plaza
PI, CNRS
serge.plaza@univ-tlse3.fr

Patrice Thuleau
Researcher, CNRS
patrie.thuleau@univ-tlse3.fr

Bertrand Fabre
Researcher, CNRS
bertrand.fabre@univ-tlse3.fr

Carine Duboe
Technical staff, CNRS
carine.duboe@univ-tlse3.fr

Clémence Guillon
PhD student
clemence.guillon@univ-tlse3.fr

Nathanael Jariais
Ingénieur
nathanael.jariais@univ-tlse3.fr
Former People
Bruno Guillotin
Amaia Nogales
Marlène Alexandre
Agustina Llanos
Madalen Le Gorrec
Patrizia Tavormina
Sabine Martin
Anne Prel
Audrey Montigny
Sabine Tourneur
Mélanie Ormancey
Many disciplines, from human genetics to plant breeding, microbiology and virology, commonly face the challenge of understanding the functions of an increasing numbers of genes. However, a major challenge has still to be accomplished in understanding what exactly the genomes code and how they function. During the two last decades, genome profiling of many species has disclosed the existence of numerous novel genes and mechanisms lacking the classical hallmarks of protein-coding genes. One striking outcome was the discovery of the small interfering RNAs twenty years ago and, more recently the long non-coding RNAs encoding (or not) micropeptides, representing one paradigm of alternative genic function contrasting with the classic coding gene model.

Our interest is centered on the study of the biology and the discovery of non-conventional regulatory mechanisms. More specifically, our objectives are centered to the understanding of the biology of microRNAs and the function of regulatory peptides in the expression of the genome. We address these questions in different model systems, such as several plants species, bacteria, fungi or drosophila.
We study the developmental functions, the underlying molecular mechanisms and their possible applicability. We concentrate at present our studies in two main directions:
– identify the molecular mechanisms involved in the regulation of expression and the activity of microRNAs (miRNAs), some of them being regulated by peptides.
– Identify and study new classes of regulating peptides.
These small molecules constitute natural active ingredients capable of modulating the expression or controlling the activity of genes. They present a potential interest as environment-friendly means of fight to favor the resistance of plants (or animals) against infectious agents. So, in parallel of the basic research, we develop a translational research when possible and maintain a narrow link with industrial companies
Identifying and studying new classes of regulatory peptides
Beyond expanding our fundamental knowledge of the mechanisms of gene regulation, miPEPs or peptides?are likely to be useful as original tools for a wide range of applications. We are investigating whether similar uses can be made of miPEPs to study miRNA regulation of gene expression in other organisms.
Moreover, the discovery of miPEPs raises the question of the complexity of the ‘peptidome‘ of an organism, especially how many and what types of peptides have functional roles in the cell. The study of miPEPs will certainly contribute answers to this question and may provide new tools to investigate the diversity and the roles of peptides.
Like miRNAs, we fully expect that peptides to be found in all plant and animal species constitute an important and ubiquitous facet of gene regulation. Demonstrating whether or not this is the case is one of the fundamental objectives of the team. If this indeed proves to be the case, peptides are likely to find applications beyond agronomy, in the control of infectious diseases and in animal and human health. Nevertheless, the fundamental concept is still in its infancy, and there is a huge amount of work needed to finely unravel the underlying biological mechanisms. This is the purpose of the team for the next years.
Ormancey M, Thuleau P, Combier JP, Plaza S. The Essentials on microRNA-Encoded Peptides from Plants to Animals. Biomolecules 13 (2), 206
Ormancey M, Guillotin B, Merret R, Camborde L, Duboé C, Fabre B, Pouzet C, Impens F, Van Haver D, Carpentier MC, San Clemente H, Aguilar M, Lauressergues D, Scharff LB, Pichereaux C, Burlet-Schiltz O, Bousquet-Antonelli C, Gevaert K, Thuleau P, Plaza S, Combier JP. Complementary peptides represent a credible alternative to agrochemicals by activating translation of targeted proteins. Nature Communications 14 (1), 254
Dozier C, Montigny A, Viladrich M, Culerrier R, Combier JP, Besson A, Plaza S. Small ORFs as New Regulators of Pri-miRNAs and miRNAs Expression in Human and Drosophila. Int J Mol Sci. 2022;23(10):5764
Fabre B, Duboé C, Pichereaux C, Montigny A, Korona D, Brun C, Camus M, Burlet-Schiltz O, Russell S, Combier JP, Lilley KS and Plaza S. In depth exploration of the alternative proteome of Drosophila melanogaster.. Frontiers Cell and Developmental Biology, 2022;10:901351.
Dozier C and Plaza S. Functions of animal microRNA-encoded peptides: the race is on! EMBO Rep. 2022:e54789.
Lauressergues D, Ormancey M, Guillotin B, San Clemente H, Duboé C, Charpentier P, Barozet A, Thuleau P, Plaza S, Combier JP. Characterization of plant microRNA-encoded peptides (miPEPs) reveals molecular mechanisms from the translation to activity and specificity. Cell Rep. 2022;38(6):110339.
Ormancey M, Guillotin B, San Clemente H, Thuleau P, Plaza S, Combier JP.Use of microRNA-encoded peptides to improve agronomic traits. Plant Biotechnol J. 2021 19(9):1687-1689.
Montigny A, Tavormina P, Duboe C, San Clémente H, Aguilar M, Valenti P, Lauressergues D, Combier JP, Plaza S. Drosophila primary microRNA-8 encodes a microRNA-encoded peptide acting in parallel of miR-8.Genome Biol. 2021;22(1):118.
Prel A, Dozier C, Combier JP, Plaza S, Besson A.Evidence That Regulation of Pri-miRNA/miRNA Expression Is Not a General Rule of miPEPs Function in Humans. Int J Mol Sci. 2021;22(7):3432.
Fabre B, Combier JP, Plaza S.Recent advances in mass spectrometry-based peptidomics workflows to identify short-open-reading-frame-encoded peptides and explore their functions. Curr Opin Chem Biol. 2021;60:122-130.
Ormancey M, Le Ru A, Duboé C, Jin H, Thuleau P, Plaza S, Combier JP. Internalization of miPEP165a into Arabidopsis Roots Depends on Both Passive Diffusion and Endocytosis-Associated Processes. Int J Mol Sci. 2020 ;21(7):2266.
Plaza S, Menschaert G, Payre F. In Search of Lost Small Peptides. Annu Rev Cell Dev Biol. 2017
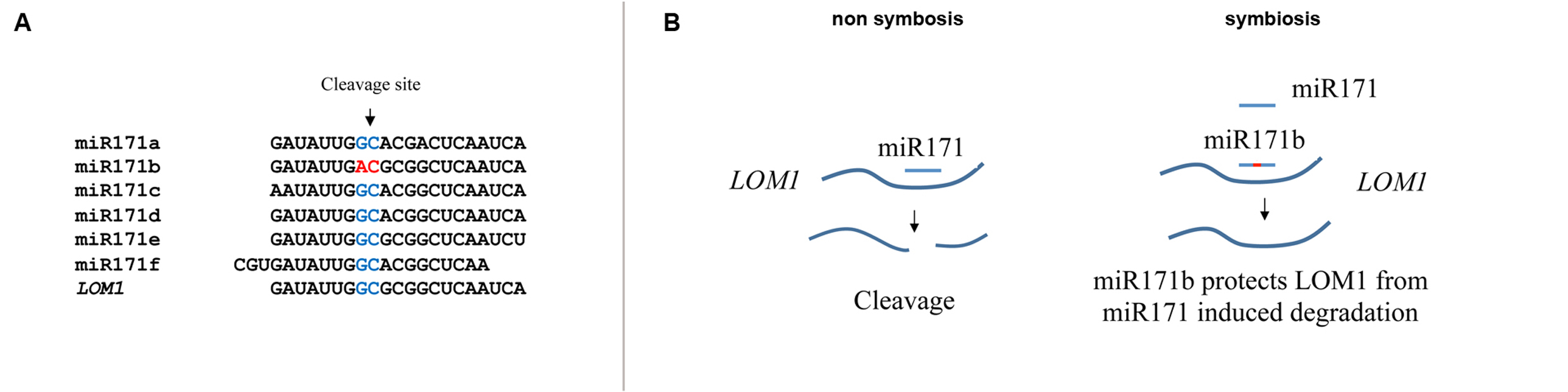
Couzigou JM, Lauressergues D, André O, Gutjahr C, Guillotin B, Bécard G, Combier JP. Positive gene regulation by a natural protective miRNA enables Arbuscular Mycorrhizal Symbiosis. Cell, Host and Microbe. 2017.
Guillotin B, Couzigou JM, Combier JP. NIN is involved in the regulation of Arbuscular Mycorrhizal symbiosis. Frontiers in Plant Science. 2016. 7:1704.
Guillotin B, Etemadi M, Audran C, Bouzayen M, Bécard G, Combier JP. Sl-IAA27 regulates strigolactone biosynthesis and mycorrhization in Tomato (var. MicroTom). New Phytologist. 2016.
Couzigou JM, Combier JP. Plant miRNAs : key regulators of root architecture and biotic interactions. New Phytologist. 2016. 212:22-35.
Zanet J, Chanut-Delalande H, Plaza S, Payre F. Small Peptides as Newcomers in the Control of Drosophila Development. Current Topics in Developmental Biology. 2016.117:199-219.
Couzigou JM, André O, Guillotin B, Alexandre M, Combier JP. Use of microRNA-encoded peptide miPEP172c to stimulate nodulation in soybean. New Phytologist. 2016. 211:379-381.
Couzigou JM, Lauressergues D, Bécard G, Combier JP. miRNA-encoded peptides (miPEPs) : a new tool to analyse the roles of miRNAs in plant biology. RNA biology. 2015. 12:1178-1180.
Lauressergues D, Couzigou JM, San Clemente H, Martinez Y, Dunand C, Bécard G, Combier JP. Primary transcripts of microRNAs encode regulatory peptides. Nature. 2015. 520:90-3.
Zanet J, Benrabah E, Li T, Pélissier-Monier A, Chanut-Delalande H, Ronsin B, Bellen H, F Payre, S Plaza. Pri sORF peptides induce selective proteasome-mediated protein processing. Science. 2015. 349(6254):1356.
lrsv.gestion@univ-tlse3.fr.
Phone
Standard : +33 5.34.32.38.01
Fax : +33 5.34.32.38.02
Find us
24, chemin de Borde-Rouge.BP 42617 Auzeville.
31326, Castanet-Tolosan. FRANCE