Notre Equipe
Christophe DUNAND
Co-leader of the team - Professor - Paul Sabatier-Toulouse 3 University
christophe.dunand@univ-tlse3.fr, https://www.researchgate.net/profile/Christophe-Dunand
Elisabeth JAMET
Co-leader of the team - Research Director CNRS
Vincent BURLAT
Professor - Paul Sabatier-Toulouse 3 University
vincent.burlat@univ-tlse3.fr, https://www.researchgate.net/profile/Vincent-Burlat

Josiane CHOURRE
Technician - Paul Sabatier-Toulouse 3 University
josiane.chourre@univ-tlse3.fr
Laurent HOFFMANN
Assistant Professor - Paul Sabatier-Toulouse 3 University
laurent.hoffmann@univ-tlse3.fr
Catherine MATHE
Assistant Professor - Paul Sabatier-Toulouse 3 University
catherine.mathe-dehais@univ-tlse3.fr

Steven MOUSSU
Researcher CNRS
steven.moussu@univ-tlse3.fr

Philippe RANOCHA
Researcher CNRS
philippe.ranocha@univ-tlse3.fr
David ROUJOL
Engineer - Paul Sabatier-Toulouse 3 University
david.roujol@univ-tlse3.fr
Denise ARICO
Post-doctoral fellow ERC Musix
denise.arico@ens-lyon.fr
Thomas BERTHELIER
PhD student - Paul Sabatier-Toulouse 3 University
thomas.berthelier@univ-tlse3.fr

Aurélie DUPRIEZ
PhD student - Paul Sabatier-Toulouse 3 University
aurelie.dupriez@univ-tlse3.fr

Sébastien CABANAC
PhD student - Paul Sabatier-Toulouse 3 University
sebastien.cabanac@univ-tlse3.fr
David VERA PINARGOTE
PhD student - Paul Sabatier-Toulouse 3 University
luis-david.vera-pinargote@univ-tlse3.fr

Mathieu HENNEUSE
Master student - Paul Sabatier-Toulouse 3 University
mathieu.henneuse@univ-tlse3.fr
Former members
Cécile ALBENNE (assistant professor UPS, 2005-2016)
Maxime BAFOIL (PhD student, 2018-2020, post-doctoral fellow, 2021-2022)
Kevin BELLANDE (PhD student, 2015-2018)
Hervé CANUT (CNRS researcher, -2023)
Maria-Juliana CALDERAN-RODRIGUES (PhD student, São Paulo University, Brasil, COFECUB, 2010, 2011)
Bastien DAUPHIN (PhD student, 2019-2022, post-doctoral fellow, 2023)
Harold DURUFLE (PhD student, 2014-2017)
Ali ELJEBBAWI (PhD student, 2018-2021)
Nizar FAWAL (PhD student, 2011-2013)
Antoine FIRMIN (PhD student, 2019-2023)
Edith FRANCOZ (PhD student/contractor assistant professor, 2012-2016)
Jonathas Pereira GRACAS (PhD student, São Paulo University, Brasil, CAPES fellowship, 2017)
May HIJAZI (PhD student, 2008-2011)
Muhammad IRSHAD (PhD student, 2005-2008)
Adélaïde JACQ (PhD student, 2013-2016)
Achraf JEMMAT (Engineer, IdEx contract, 2015-2016)
Hasan KOLKAS (PhD student, 2019-2022)
Qiang LI (PhD student, 2010-2014)
Duchesse MBADINGA MBADINGA (PhD student, 2017-2020)
Huan NGUYEN-KIM (PhD student, 2011-2015)
Valérie PACQUIT (assistant professor UPS, 2009-2020)
Artur PINSKI (PhD student, Silesia University in Katowice, Poland, ERASMUS+ fellowship, 2020)
Sébastien VIUDES (PhD student, 2018-2021)
Rafael PONT-LEZICA (emeritus Professor UPS, -2011)
Research Topics
For a long time considered as a rigid envelopes surrounding plant cells, the cell walls now appears as a dynamic compartments involved in many physiological processes such as developpement, signaling or defense against biotic and abiotic stresses. Plant cell walls are mainly composed of polysaccharides (about 90% in mass) but they also contain proteins, forming altogether complex supramolecular scaffolds. Their biochemical composition and the organization of their components change during development and in response to environmental factors. The proteins play key roles in the remodeling of the cell wall architecture, by modifying the polysaccharides, generating signals or interacting with the other cell wall components.
Our work aims at the understanding of the roles of the cell wall proteins during plant development. It is then necessary (i) to identify the whole cell wall protein complement, (ii) to follow the dynamics of the proteome in different physiological conditions, and (iii) to perform functional studies. These functional studies have led to the description of cell wall microdomains performing specific functions during particular developmental stages and of non covalent proteins/polysaccharides networks possibly allowing a quick reorganization of the cell wall networks during rapid growth phases. The reactive oxygen species (ROS), which are key players of many physiological processes, are also involved in the cell wall dynamics. We look at the control of their production by cell wall proteins as well as their localization. Finally, we are interested in the evolution of some cell wall protein families by studying different organisms along the green lineage, from aquatic plants considered as ancestors of land plants, to plants appeared more recently during its evolution.
Our team has also developed several bioinformatic tools open to the scientific community. Three databases have been created and are regularly updated : Redoxibase, ProtAnnDB and WallProtDB.
- Cell wall proteomics
- Cell wall architecture
- Evolution of cell wall proteins
- Cell wall plasticity and adaptation
- Signaling
Cell wall proteomics
Contacts: Elisabeth JAMET, Laurent HOFFMANN
PhD students: Muhammad IRSHAD (2005-2008), Huan NGUYEN-KIM (2011-2015), Harold DURUFLE (2014-2017), Hasan KOLKAS (2019-2022)
Master students: Leïla FEIZ (2005), Yu ZHANG (2009), Aurélie GIBOULOT (2010), Kahina MERAH (2012), Vincent HERVE (2014),
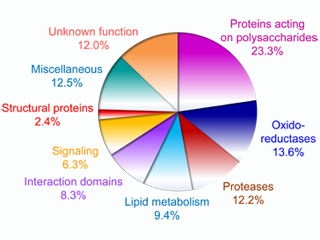
Since the beginning of the 2000’s, our team has performed plant cell wall proteomics studies and has acquired an international reputation in this area. We have greatly contributed to the analysis of the cell wall proteome of the model plant Arabidopsis thaliana which represents the best described cell wall proteome to date with 975 identified proteins (Albenne et al. 2013). Several materials have been studied in our team: cell suspension cultures (Borderies et al. 2003), roots (Nguyen-Kim et al. 2016), rosettes (Boudart et al. 2005, Hervé et al. 2016), stems (Duruflé et al. 2017), the secretome of etiolated plantlets cultivated in liquid medium (Charmont et al. 2005) or etiolated hypocotyls (Feiz et al. 2006, Irshad et al. 2008).
Protocols dedicated to the purification of cell walls (Feiz et al. 2006) and to the extraction of cell wall proteins from the purified cell walls with salt solutions (Irshad et al. 2008) have been established. They have allowed the analysis of the cell wall proteomes of different plant sepcies: Brachypodium distachyon (Douché et al. 2013, 2021; Francin-Allami et al. 2015, 2016; Pinski et al. 2021), Saccharum officinarum (Calderan-Rodrigues et al. 2014, 2016, 2017; Fonseca et al. 2018) and Triticum aestivum (Cherkaoui et al. 2018, 2020). This work has shown the great diversity of cell wall proteins as well as the differences between the proteomes of different organs, thus illustrating the cell wall plasticity.

Beyond the identification of the proteins, we have described post-translational modifications like N-glycosylations (Ligat et al. 2011, Zhang et al. 2011), hydroxylation of proline residues (Pro) in hydroxyprolines (Hyp) (Nguyen-Kim et al. 2016, Duruflé et al. 2017) or O-glycosylation of Hyp residues (Hijazi et al. 2012). We have been able to propose a new code to predict the occurrence of Hyp residues in the amino acid sequences of the hydroxyproline-rich glycoproteins (HRGPs) (Canut et al. 2016, Duruflé et al. 2017).
The ProtAnnDB database has been created to annotate the proteins identified by mass spectrometry and bioinformatics on the basis of the prediction of sub-cellular localization and of functional domains annotated by experts (San Clemente et al. 2009). This annotation allows comparing the different proteomes collected in the WallProtDB database (San Clemente and Jamet 2015). This work has allowed distinguishing two groups of proteins: (i) those which have a predicted signal peptide and which are called cell wall proteins, and (ii) those which have no predicted signal peptide and could be either intracellular contaminants or secreted by alternative pathways yet to be described in plants (Albenne et al. 2013, Regente et al. 2017). More recently, the mass spectrometry data have been used to compare the N- and C-terminal ends of the cell wall proteins determined experimentally to those predicted by bioinformatics (Pinski et al. 2021).
We have contributed to the creation of new cloning vectors to quickly check the sub-cellular localization of proteins of interest identified in the cell wall proteomes. We can easily obtain proteins in fusion with different kinds of fluorescent proteins which are stable at the acidic pH of the cell walls, like the TagRFP, the TagBFP or the T-Sapphire (Albenne et al. 2014; Berthold, Roujol et al. 2019; Roujol et al. 2020; Pinski et al. 2021).
Our studies are now focused on the study of the cell wall proteomes of plants considered as ancestors of the green lineage as well as of other plant species through collaborations. It should allow us to study the evolution of the plant cell wall proteome. Besides, we are involved in an ANR project called STRESS-PEPT which is devoted to the study of the role of extracellular peptides during plant-microorganisms interactions.
Collaborations:
– Mathilde FRANCIN-ALLAMI, Fabienne GUILLON, Colette LARRE, Mehdi CHERKAOUI, Biopolymères Interactions Assemblages, INRAE Nantes.
– Catherine RAYON, Karine PAGEAU, Maïté LESCHEVIN, Biologie des Plantes et Innovation, Picardie-Jules Verne University, Amiens.
– Sébastien AUBOURG, Jean-Pierre RENOU, Philippe GRAPPIN, Institut de Recherche en Horticulture et Semences, INRAE Angers, Coordination ANR STRESS-PEPT.
– Michel ZIVY, Thierry BALLIAU, Plateforme d’analyse protéomique Paris Sud-Ouest, Le Moulon.
– Carlos LABATE, Maria-Juliana CALDERAN-RODRIGUES, Laboratório Max Feffer de Genética de Plantas, Escola Superior de Agricultura “Luiz de Queiroz”, São Paulo University, Piracicaba, Brasil)
– Laura de la CANAL, Instituto de Investigaciones Biológicas, Universidad Nacional de Mar del Plata, Argentina.
– Alexander BETEKHTIN, Artur PINSKI, Faculty of Natural Sciences, University of Silesia in Katowice, Poland.
– Steven FRY, University of Edinburgh , UK.
1. Dynamics of cell wall microdomains during development and seed germination in Arabidopsis thaliana
Contacts: Vincent BURLAT, Christophe DUNAND, Philippe RANOCHA
PhD students: Edith FRANCOZ (2012-2015), Sébastien VIUDES (2018-2021), Bastien DAUPHIN (2019-2022)
Plant cell walls are highly dynamic structures that are constantly remodeled during plant development. We are particularly interested in the molecular scaffolds enabling either loosening or stiffening events of cell wall microdomains which are necessary for proper establishment of cellular architecture, in turn necessary for proper plant development. We mainly use two Arabidopsis thaliana cellular models to tackle this question: (i) the seed mucilage secretory cells (Francoz et al. 2015, Francoz et al. 2019) and (ii) the seed micropylar endosperm (Jemmat et al. 2020). The main molecular actors that we are interested in are class III peroxidases (CIII-Prxs) that are able to either loosen or stiffen cell wall microdomains depending on the considered isoform within the 73 members of the multigenic family (Francoz et al. 2015). Following an in situ hybridization screening step of spatiotemporal expression profiles of CIII Prxs genes during A. thaliana seed development (Francoz et al. 2016) and seed germination (Jemmat et al., 2020), we have identified candidate genes for further functional studies. We extensively studied the molecular mechanism enabling the positioning the CIII Prx PRX36 to a mucilage secretory cell wall microdomain during seed development. The proper release of mucilage during seed imbibition necessitates the loosening of a mucilage secretory cell wall microdomain earlier during seed development. Using an integrated genetics-cell biology-molecular modelling study, we have discovered (i) the occurrence within this cell wall microdomain of a partially demethylesterified homogalacturonan molecular pattern controlled by PECTIN METHYLESTERASE INHIBITOR 6 (PMEI6), which constitutes (ii) an anchoring molecular platform for PRX36, allowing (iii) its highly polarized loosening action necessary for proper mucilage release (Francoz et al. 2019).
We currently pursue the study of this concept of molecular scaffolds enabling the anchoring of CIII-Prxs on homogalacturonan barcodes in the frame of the ANR-funded collaborative research project MicroWall that we coordinate.
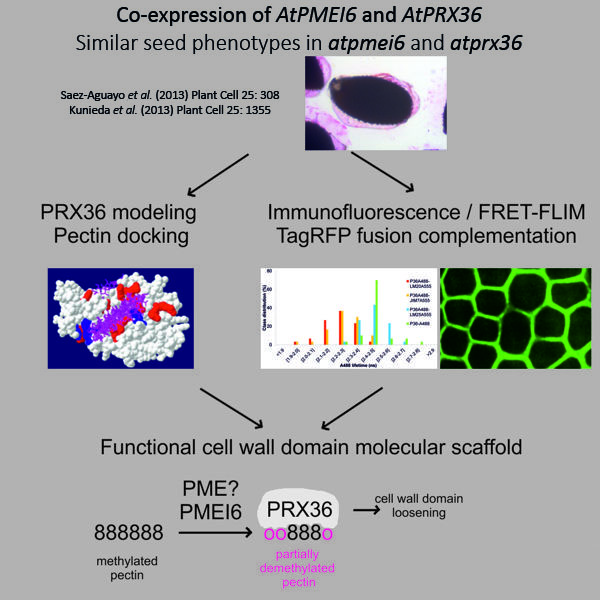
Collaborations:
– Jérôme PELLOUX, Biologie des Plantes et Innovation, Picardie-Jules Verne University, Amiens, Partner ANR MicroWall.
– Marie-Christine RALET, Biopolymères Interactions Assemblages, INRAE Nantes, Partner ANR MicroWall.
2. Supra-molecular architecture in plant cell walls: evidence for the existence of non-covalent proteins/polysaccharides networks
Contact: Elisabeth JAMET
PhD students: Muhammad IRSHAD (2005-2008), May HIJAZI (2008-2011), Huan NGUYEN-KIM (2011-2015), Hasan KOLKAS (2019-2022)
During growth, plant cell walls have to be modified to allow the increase in size of the cells. We have focused our work on AGP31 (ArabinoGalactan Protein 31), a protein of Arabidopsis thaliana encoded by At1g28290, because it has been identified as a major protein in the cell wall proteome of etiolated hypocotyls, which cells undergo a great elongation within a short time (Irshad et al. 2008). AGP31 has a N-terminal signal peptide, an arabinogalactan protein (AGP) domain, a His-rich region, a central Pro-rich domain and a C-terminal domain called PAC (Proline-rich protein, Arabinogalactan protein, conserved Cys). Different mass spectrometry and biochemistry approaches have allowed showing the presence of hydroxyproline (Hyp) residues and of Gal/Ara-rich O-glycans in the Pro-rich domain (Hijazi et al. 2012). AGP31 has been found as various glycoforms differing by the type and the size of the O-glycans and/or the length of the polypeptide chain. Besides, interactions between the AGP31 PAC domain and the galactans present on type I rhamnogalacturonans (RG I) have been shown in vitro (Hijazi et al. 2014). AGP31 has also been shown to interact with itself, most probably through the O-glycans of its Pro/Hyp-rich domain and its PAC domain. Finally, a remarkable affinity has been found between these O-glycans and a peanut lectin (PNA, PeaNut Agglutinin), suggesting interactions with cell wall lectins (Hijazi et al. 2012).
All these data have allowed proposing that AGP31 is the center of non-covalent supramolecular scaffolds, thus playing a structural role in cell walls. They bring new concepts regarding the architecture of cell walls and highlight the central role of the O-glycoproteins in their organization (Hijazi et al. 2014).

The role of the AGP31 PAC domain in these supramolecular scaffolds has prompted us to look for the presence of PAC domains in other cell wall proteins. We have been able to show that PAC domain proteins are present in plants considered as ancestors of the flowering plants, like Marchantia polymorpha, as well as in all the plants studied so far (Nguyen-Kim et al. 2020).
The present studies aim at understanding the evolution of the PAC domain from structural and functional point of views.
Collaboration: Gabriele GADERMAIER, Josef LAIMER, Peter LACKNER, Salzbourg University, Austria.
1. Repertoire of genes involved in the regulation of reactive oxygen species (ROS): the RedoxiBase
Contacts: Catherine MATHÉ, Christophe DUNAND, Bruno SAVELLI
PhD students: Qiang LI (2011-2015), Nizar FAWAL (2012-2015)
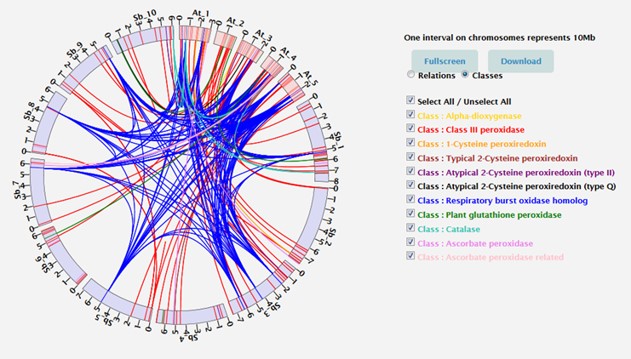
La Redoxibase centralise des séquences codant pour des oxydo-réductases de tous les organismes vivants. L’annotation manuelle et la correction de séquences déjà annotées sont nos préoccupations majeures, mais aussi une garantie de qualité. Cependant, avec le nombre croissant d’organismes et de séquences disponibles, la RedoxiBase évolue régulièrement pour rester compétitive et attractive (Savelli et al. 2019).
Le projet a pour objectifs : (i) de mettre en place des pipelines afin d’accélérer l’intégration de nouvelles oxydo-réductases, en vérifiant et corrigeant automatiquement les peroxydases annotées lors des nombreux projets de séquençage génomique. Pour cela, les séquences sont soumises à nos profils familles-spécifiques et comparées en terme de structure exon/intron aux séquences de la base ; (ii) de développer de nouveaux outils mis à disposition via l’interface de la RedoxiBase afin de faciliter l’analyse comparative intra-espèces et inter-espèces : visualisation des polymorphismes, outils de cartographie ou de navigation sur les génomes (e.g. MapChart, GBrowse), identification et représentation des relations d’orthologie, paralogie (e.g. SyMap, Circos).
2. Search for class III peroxidases (CIII Prxs) and Study of their evolution in the green lineage
Contacts: Catherine MATHÉ, Christophe DUNAND
PhD student: Duchesse MBADINGA MBADINGA (2017-2020)
Peroxidases catalyze redox reactions involving hydrogen peroxide and a variable substrate. These proteins are present in all kingdoms and generally form multigenic families of variable size (from 2 to 138 isoforms). The non-animal peroxidase superfamily contains three classes (I, II and III) with closely related tertiary structures and probably sharing a common ancestral sequence. Class I and III peroxidases (CI and CIII Prxs) are found in plants (Mbadinga Mbadinga et al. 2020). They have fundamental roles in different physiological processes such as reactive oxygen species (ROS) detoxification, defense against pathogens or cell wall formation (Cosio et al. 2009). CI and CIII Prx genes show a high level of duplications in plants.
The main objectives of the projectare as follows: (i) to obtain a very precise picture of the taxonomic distribution of CIII Prx genes within Charaphyceae; (ii) to reconstruct ancestral sequences on the main taxonomic nodes.

Cell wall plasticity and adaptation
Contacts: Christophe DUNAND, Vincent BURLAT, Philippe RANOCHA
PhD students: Ali ELJEBBAWI (2018-2021), Sébastien VIUDES (2018-2021)
In Arabidopsis thaliana, the 73 class III peroxidases (CIII Prxs) show specific expression profiles (Francoz et al., 2015). Recent studies tend to show that the fine localization of individual CIII Prxs could lead to a specific “enzyme-substrate” encounter allowing a controlled action in particular cell wall micro-domains. This action could contribute to cell wall loosening or stiffening.
The developmental processes leading to seed formation and seed germination were chosen to study the role of CIII Prxs. In A. thaliana, the cells located in the outermost epidermal layer of the seed (mucilage-secretory cells, MSCs) synthesize a volcano-shaped inner wall and produce abundant compressed polysaccharidic mucilage. The amount, form or composition of mucilage varies widely within Brassicaceae.
The rupture of the seed coat (testa and micropylar endosperm) is critical for germination by allowing the emergence of the radicle. On the other hand, we have shown that the production of reactive oxygen species (ROS) is necessary for germination, and that CIII Prxs could be involved in ROS homeostasis.
The main objectives of the project are as follows: (i) to better understand the evolution of MSC establishment, mucilage production and extrusion mechanisms within Brassicaceae; (ii) to analyze several CIII Prx candidates that may be involved in the control of micropylar endosperm disruption.

1. Reactive oxygen species (ROS) homeostasy during root development
Contacts: Christophe DUNAND, Philippe RANOCHA
PhD student: Ali ELJEBBAWI (2018-2021)
Reactive oxygen species (ROS), well known for their deleterious effects during stress and defense responses, may also have signaling roles. The control of ROS homeostasis during biological processes such as root development is essential. This includes cell elongation, the development of root hairs and the appearance of secondary roots. Three transcription factors each controlling the expression of distinct class III peroxidases (CIII Prxs), whose general biological function is to control ROS homeostasis, were studied in Arabidopsis thaliana (Eljabbawi et al. 2021).
The main objectives of the projectare as follows: (i) the functional analysis of three CIII Prxs from A. thaliana during root hair development under contrasting growth conditions; (ii) the identification of new players related to the control of root architecture and homeostasis of ROS in response to contrasting growing conditions, such as temperature and saline stresses and their combined effects. For this, the nuclear genome of several individuals from different populations of A. thaliana rae presently sequenced and high throughput phenotypic analysis on the root system of each cultured ecotype under three temperature conditions (16°C, 22°C, 28°C) and two salt conditions (0 mM and 50 mM) are performed.

Collaboration: José Manuel ESTEVEZ, Fundación Instituto Leloir, Buenos Aires, Argentina.
2. Lectin-receptor kinases and Perception of the cell wall integrity
Contact: Hervé CANUT
PhD students: Anne GOUGET (2002-2006), Kevin BELLANDE (2016-2019)
Our team is interested in the communication between the cell wall and the cytoplasm of plant cells. The project aims at understanding the roles of lectins in the model plant Arabidopsis thaliana.
Lectins are present in all the living world. They are able to interact with mono- or oligo-saccharides in a reversible way and to decode the information carried by these complex structures. Such molecules, mostly originating from plants, have found many applications in biological sciences and in medicine. Since their first use for blood typing, their role, often beneficial, has been demonstrated on the development of tumors and in therapy against cancer. In the same way, lectins are essential for plants and play many roles in cell-to-cell communication, development and defense reactions.
Plant cell walls are complex structures containing cellulose, hemicelluloses, pectins and proteins secreted by the cells, thus creating an extracellular matrix connecting cells in tissues. Cell walls are dynamics and sources of mechanical and chemical signals. In A. thaliana, lectins are present in cell walls as soluble proteins weakly interacting with cell wall components (Lecs) or as extracellular domaines of lectin receptor kinases (lecRKs) (Bellande et al. 2017).
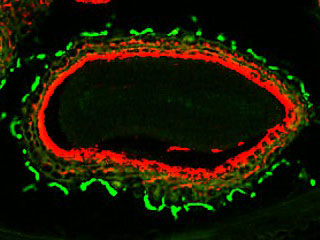
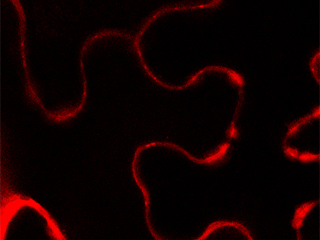
We have shown that the plasma membrane-cell wall contacts [1-white arrow] are established by protein-protein interactions involving the recognition of the Arg-Gly-Asp (RGD) motif in A. thaliana. LecRK-I.9 (At5g60300) is considered as a putative link between the plasma membrane and the cell wall (Gouget et al. 2006). It belongs to the legume lectin family [2]. We have shown that the resistance to Pseudomonas syringae pv tomato (Pst), a pathogen bacteria, and to Phytophthora infestans, an oomycete, is impaired in a lecrk-I.9 mutant (Bouwmeester et al. 2011, Balagué et al. 2016).
Besides, our analysis of the cell wall proteome of elongating cells has identified several lectins (Irshad et al. 2008). Phylogenic analyses have shown that the Lecs and the extracellular domains of LecRKs are very close to each other and form a cluster (Bellande et al. 2017). Finally, Enfin, “LecRK promoter-I.9::GUS” constructs have shown that the activity of the LecRK-I.9 promoter is the highest in the main root apex and is absent from lateral root primordia [3].

The aim of this project is to understand the role of Lecs and LecRKs. Several aproaches, including biochemitry and functional genomics, are performed to identify the extracellular ligands of these proteins. We assume that they are involved in the perception of pathogens, but also in the assembly and the remodeling of cell wall polysaccharides. Lecs and LecRKs could act as sensors of cell wall integrity.
ANR WALLARRAY (2009)
ANR WALLARRAY2 (2011-2014)
Collaborations:
Francine GOVERS, Laboratory of Phytopathology, Wageningen University, The Netherlands.
Olivier HAMANT, Laboratoire de Reproduction et Développement des plantes, Lyon. MUSIX ERC project (2021-2025).
The team has organized:
- The French Plant cell wall Network Days (2008).

- The annual meeting of the Green Proteome (2011).

- The mixOmics Summer School in the frame of COST FA1306 The quest for tolerant varieties : phenotyping at plant and cellular level (2016).
Elisabeth JAMET has been invited as a co-editor together with Els VAN DAMME, Nausicaa LANNOO and Cécile ALBENNE of an e-book dealing with Plant Glycobiology – A sweet world of lectins, glycoproteins, glycolipids and glycans of a Research Topics of Frontiers in Plant Science (2015) and invited co-editor together with Véronique SANTONI of a Special issue of Proteomes entitled Plant Proteomics 2017. She has also been a co-editor (i) with Christophe DUNAND of a Special Issue of International Journal of Molecular Sciences entitled Plant Cell wall Proteins and Development, (ii) with C DUNAND and Zoë POPPER of a Special Topic in Frontiers in Plant Science dealing with Co-evolution of plant cell wall polymers and (iii) with C DUNAND of a Special Issue of Cells entitled Research on Plant cell Wall Biology.
C DUNAND is presently editing a Topical Collection in International Journal of Molecular Sciences entitled The plant cell walls and their impact on plant physiology. He is also co-editing with Steven MOUSSU a Special Issue of International Journal of Molecular Sciences entitled ROS in plant cell polar growth. These two Special Issues are presently opened for submission.
Publications
2024
2023
Arico DS, Dickmann JEM, Hamant O, Canut H (2023) The plasma membrane – cell wall nexus in plant cells: focus on the Hechtian structure. Cell Surf 10: 100115 (read)
Badruna L, Burlat V, Montanier CY (2023) CBMs as probes to explore plant cell wall heterogeneity using immunocytochemistry. Methods Mol Biol 2657:163-179 (read)
Burlat, V, Papon N, Courdavault V (2023) Medicinal plants enter the single-cell multi-omics era. Trends Plant Sci 28: 1205-1207 (read)
Jamet E, Esquerré-Tugayé M-T, Gallardo-Guerrero K, Rolland N, Zivy M, Blein-Nicolas, Vincent D, Gontero B, Rajjou L (2023) Obituary: Dominique Job (1947-2022). Front Plant Sci 14: 1188766 (read)
Jamet E, Dunand C (2023) Plant Cell Wall Proteins and Development. MDPI (Basel). 342 pp. (read)
Kolkas H, Burlat V, Jamet E (2023) Immunochemical identification of the main cell wall polysaccharides of the early land plant Marchantia polymorpha. Cells 12: 1833 (read)
Moussu S, Ingram G (2023) The EXTENSIN enigma. Cell Surf 9: 10094 (read)
2022
2021
Althiab-Almasud R, Chen Y, Maza E, Djari A, Frasse P, Mollet JC, Mazars C, Jamet E, Chervin C (2021) Ethylene signaling modulates tomato pollen tube growth, through modifications of cell wall remodeling and calcium gradient. Plant J 107: 893-908 (read)
Badruna L, Burlat V, Roblin P, Enjalbert T, Lippens G, Venditto I, O’Donohue MJ, Montanier C (2021) The Jo-In protein welding system is a relevant tool to create CBM-containing plant cell wall degrading enzymes. New Biotechnol 65: 31-41 (read)
Bafoil M, Yousfi M, Dunand C, Nerbahi N (2021) Effect of dielectric barrier ambient air plasma on two Brassicaceae seeds: Arabidopsis thaliana and Camelina sativa. Int J Mol Sci 22: 9923 (read)
Douché T, Valot B, Balliau T, Zivy M, San Clemente H, Jamet E (2021). Cell wall proteomic datasets of stems and leaves of Brachypodium distachyon. Data Brief 35: 106818 (read)
Duruflé H, Selmani M, Ranocha P, Jamet E, Dunand C, Déjean S (2021) A powerful framework for an integrative study with heterogeneous omics data: from univariate statistics to multi-block analysis. Brief Bioinformatics 20: bbaa166 (read)
Eljebbawi A, Guerrero YDCR, Dunand C, Estevez JM (2021) Highlighting reactive oxygen species as multitaskers in root development. iScience. 24: 101978 (read)
2020
Bellande K, Lalo A, Ligat L, Roujol D, Jamet E, Canut H (2020) Recombinant N-glycosylation isoforms of Legume lectins: production and purification from Nicotiana benthamiana leaves following RuBisCO depletion. Plant Physiol Biochem 157: 441-452 (read)
Cherkaoui M, Lollier V, Geairon A, Bouder A, Larré C, Rogniaux H, Jamet E, Guillon F, Francin-Allami M (2020) Cell wall proteome of wheat grain endosperm and outer layers at two key stages of early development. Int J Mol Sci 21: 239 (read)
Duruflé H, Ranocha P, Balliau T, Zivy M, Albenne C, Burlat V, Déjean S, Jamet E, Dunand C (2020) An integrative study showing the adaptation to sub-optimal growth conditions of natural populations of Arabidopsis thaliana: a focus on cell wall changes. Cells 9: 2249 (read)
Roujol D, Hoffmann L, San Clemente H, Ritzenthaler C, Burlat V, Jamet E (2020) Plant cell wall proteomes : bioinformatics and cell biology tools to assess the bona fide cell wall localization of proteins. Methods Mol Biol 2149: 443-462 (read)
Viudes S, Burlat V, Dunand C (2020) Seed mucilage evolution: diverse molecular mechanisms generate versatile ecological functions for particular environments. Plant Cell Environ 43: 2857-2870 (read)
2019
Bafoil M, Le Ru A, Merbahi N, Eichwald O, Dunand C, Yousfi M (2019) New insights of low-temperature plasma effects on germination of three genotypes of Arabidopsis thaliana seeds under osmotic and saline stresses. Sci Rep 9: 8649 (read)
Berthold F, Roujol D, Hemmer C, Jamet E, Ritzenthaler C, Hoffmann L, Schmitt-Keichinger C (2019) Inside or outside? A new collection of Gateway vectors allowing plant protein subcellular localization or over-expression. Plasmid 105: 102436 (read)
Delaux PM, Hetherington AJ, Coudert Y, Delwiche C, Dunand C, Gould S, Kenrick P, Li FW, Philippe H, Rensing SA, Rich M, Strullu-Derrien C, de Vries J (2019) Reconstructing trait evolution in plant evo-devo studies. Curr Biol 29: R1110-R1118 (read)
Duruflé H, Albenne C, Jamet E, Dunand C (2019) Phenotyping and cell wall polysaccharide composition of five Arabidopsis ecotypes grown at optimal or sub-optimal temperatures. Data Brief 25: 104318 (read).
Duruflé H, Ranocha P, Balliau T, Dunand C, Jamet E (2019) Transcriptomic and cell wall proteomic datasets of rosettes and floral stems from five Arabidopsis thaliana ecotypes grown at sub-optimal temperature. Data Brief 27: 104581 (read)
Duruflé H, Ranocha P, Mbadinga Mbadinga DL, Déjean S, Bonhomme M, San Clemente, Viudes S, Eljebbawi A, Delorme-Hinoux V, Sáez-Vásquez J, Reichheld J-P, Escaravage N, Burrus M, Dunand C (2019) Phenotypic variation as a response to altitude-related constraints in Arabidopsis populations. Front Plant Sci 10: 430 (read)
Francoz E, Ranocha P, Dunand C, Burlat V (2019) Medium-throughput RNA in situ hybridization of serial sections from paraffin-embedded tissue microarrays. Methods Mol Biol, 1933: 99-130 (read)
Francoz E, Ranocha P, Le Ru A, Martinez Y, Fourquaux I, Jauneau A, Dunand C, Burlat (2019) Pectin demethylesterification generates platforms that anchor peroxidases to remodel plant cell wall domains. Dev Cell 48: 261-276 (read)
Mathé C, Fawal N, Roux C, Dunand C (2019) In silico definition of new lignin peroxidase classes in fungi and putative relation to fungal life style. Sci Rep. 9: 20373 (read)
Savelli B, Li Q, Webber M, Jemmat AM, Robitaille A, Zamocky M, Mathé C, Dunand C (2019) RedoxiBase: a new database for ROS homeostasis regulated proteins. Redox Biol 26: 101247 (read)
Savelli B, Picard S, Roux C, Dunand C (2019) ExpressWeb: A Web application for clustering and visualization of expression data. bioRxiv (read)
2018
Bafoil M, Jemmat AM, Martinez Y, Merbahi N, Eichwald O, Dunand C, Yousfi M (2018) Effects of low temperature plasmas and plasma activated waters on Arabidopsis thaliana germination and growth. PLoS One 13: e0195512 (read)
Calderan-Rodrigues MJ, Fonseca JG, San Clemente H, Labate CA, Jamet E (2018) Glycoside hydrolases in plant cell wall proteomes: predicting functions that could be relevant for improving biomass transformation processes. Advances in Biofuels and Bioenergy (M Nageshwara-Rao and J Soneji eds), InTechOpen access Publisher, Croatia, pp 165-182 (read)
Cherkaoui M, Geairon A, Lollier V, San Clemente H, Larré C, Rogniaux H, Jamet E, Guillon F, Francin-Allami M (2018) Cell wall proteome investigation of bread wheat (Triticum aestivum) developing grain in endosperm and outer layers. Proteomics 18: 1800286 (read)
Fonseca JG, Calderan-Rodrigues MJ, de Moraes FE, Cataldi TR, Jamet E, Labate CA (2018) Cell wall proteome of sugarcane young and mature leaves and stems. Proteomics 18: 1700129 (read)
Jacq A, Burlat V, Jamet E (2018) Plant cell wall proteomics as a strategy to reveal candidate proteins involved in extracellular lipid metabolism. Curr Protein Pept Sci 19: 190-199 (lire)
Jamet E, Santoni V (2018) Editorial for Special Issue: 2017 Plant Proteomics. Proteomes 6: 28 (read)
Nishiyama T et al. (2018) The Chara genome: secondary complexity and implications for plant terrestrialization. Cell 174: 448-464 (read)
2017
Badruna L, Burlat V, Montanier C (2017) CBMs as probes to explore plant cell wall heterogeneity using immunocytochemistry. Methods Mol Biol 1588 : 181-197 (read)
Balagué C, Gouget A, Bouchez O, Souriac C, Haget N, Boutet-Mercey S, Govers F, Roby D, Canut H (2017) The Arabidopsis thaliana lectin receptor kinase LecRK-I.9 is required for full resistance to Pseudomonas syringae and affects jasmonate signalling. Mol Plant Pathol 18 : 937-948 (read)
Bellande K, Bono JJ, Savelli B, Jamet E, Canut H (2017) Plant lectins and lectin receptor-like kinases : How do they sense the outside ? Int J Mol Sci 18 : 1164 (read)
Calderan-Rodrigues MJ, Fonseca JG, Labate CA, Jamet E (2017) Cell wall proteomics as a means to identify target genes to improve second generation biofuel production. In Frontiers in Bioenergy and Biofuels (E Jacob-Lopes and L Queiroz Zepka ed), InTechOpen access Publisher, Croatia, pp 5-24 (read)
Canut H, Albenne C, Jamet E (2017) Isolation of the cell wall. Methods Mol Biol 1511 : 171-185 (read)
Cosio C, Ranocha P, Francoz E, Burlat V, Zheng Y, Perry SE, Ripoll JJ, Yanofsky M, Dunand C (2017) The class III peroxidase PRX17 is a direct target of the MADS-box transcription factor AGAMOUS-LIKE15 (AGL15) and participates in lignified tissue formation. New Phytol 213 : 250-263 (read)
Duruflé H, Hervé V, Balliau T, Zivy M, Dunand C, Jamet E (2017) Proline hydroxylation in cell wall proteins : is it yet possible to define rules ? Front Plant Sci 8: 1802 (lire)
Duruflé H, Hervé V, Ranocha P, Balliau T, Zivy M, Chourré J, San Clemente, H, Burlat V, Albenne C, Déjean S, Jamet E, Dunand C (2017) Cell wall modifications of two Arabidopsis thaliana ecotypes, Col and Sha, in response to sub-optimal growth conditions : an integrative study. Plant Sci 263 : 183-193 (read)
Duruflé H, San Clemente H, Balliau T, Zivy M, Dunand C, Jamet E (2017) Cell wall proteome analysis of Arabidopsis thaliana mature stems. Proteomics 17, 8, 1600449 (read)
Jacq A, Pernot C, Martinez Y, Domergue F, Payré B, Jamet E, Burlat V, Pacquit V (2017) The Arabidopsis Lipid Transfer Protein 2 (AtLTP2) is involved in cuticle-cell wall interface integrity and in etiolated hypocotyl permeability. Front Plant Sci 8 : 263 (read)
Jamet E (2017) Plant cell wall proteomics : An assessment twenty years after launching. Bioinform. Proteomics Open Access J 1: 000107 (read)
Mangano S, Denita-Juarez SP, Choi HS, Marzol E, Hwang Y, Ranocha P, Melina Velasquez S, Borassi C, Barberini ML, Aptekmann AA, Muschietti JP, Nadra AD, Dunand C, Cho HT, Estevez JM (2017) The molecular link between auxin and ROS-mediated polar growth. Proc Natl Acad Sci USA 114: 5289-5294 (read)
Regente M, Pinedo M, San Clemente H, Balliau T, Jamet E, de la Canal L (2017) Plant extracellular vesicles are incorporated by cells of a fungal pathogen and inhibit its growth. J Exp Bot 20: 5485-5496 (read)
Sibout R, Proost S, Hansen B, Vaid N, Giorgi F, Ho-Yue-Kuang S, Legée F, Cezart L, Bouchabké-Coussa O, Soulhat C, Provart N, Pasha A, Le Bris P, Roujol D, Höfte H, Jamet E, Lapierre C, Persson S, Mutwil M (2017) Expression atlas and comparative co-expression network analyses reveal important genes involved in the formation of lignified cell wall in Brachypodium distachyon. New Phytol 215 : 1009-1025 (read)
2016
Calderan-Rodrigues MJ, Jamet E, Douché T, Rodrigues Bonassi MB, Regiani Cataldi T, Fonseca Guimarães J, San Clemente H, Pont-Lezica R, Labate CA (2016) Cell wall proteome of sugarcane stems : comparison of a destructive and a non-destructive extraction method showed differences in glycoside hydrolases and peroxidases. BMC Plant Biol 16 : 14 (read)
Canut H, Albenne C, Jamet E (2016) Post-translational modifications of plant cell wall proteins and peptides : A survey from a proteomics point of view. Biochim Biophys Acta 1864 : 983-990 (read)
Francin-Allami M, Lollier V, Pavlovic M, San Clemente H, Rogniaux H, Jamet E, Guillon F, Larré C (2016) Understanding the remodelling of cell walls during Brachypodium distachyon grain development through a sub-cellular quantitative proteomic approach. Proteomes 4 : 21 (read)
Francoz E, Ranocha P, Pernot C, Le Ru, A, Pacquit V, Dunand C, Burlat V (2016) Complementarity of medium-throughput in situ RNA hybridization and tissue-specific transcriptomics : case study of Arabidopsis seed development kinetics. Sci Rep 6 : 24644 (read)
Hervé V, Duruflé H, San Clemente H, Albenne C, Balliau T, Zivy M, Dunand C, Jamet E (2016) An enlarged cell wall proteome of Arabidopsis thaliana rosettes. Proteomics 16 : 3183-3187 (read)
Nguyen-Kim H, San Clemente H, Balliau T, Zivy M, Dunand C, Albenne C, Jamet E (2016) Arabidopsis thaliana root cell wall proteomics : increasing the proteome coverage using a combinatorial peptide ligand library and description of unexpected Hyp in peroxidase amino acid sequences. Proteomics 16 : 491-503 (read)
2015
Cao PB, Azar S, San Clemente H, Mounet F, Dunand C, Marque G, Marque C, Teulières C (2015) Genome-wide analysis of the AP2/ERF family in Eucalyptus grandis: an intriguing over-representation of DREB1/CBF genes all stress responsive. PLoS One 10: e0121041 (read)
Delaux PM, Radhakrishnan GV, Jayaraman D, Cheema J, Malbreil M, Volkening JD, Sekimoto H, Nishiyama T, Melkonian M, Pokorny L, Rothfels CJ, Sederoff HW, Stevenson DW, Surek B, Zhang Y, Sussman MR, Dunand C, Morris RJ, Roux C, Wong GK, Oldroyd GE, Ané JM (2015) Algal ancestor of land plants was preadapted for symbiosis. Proc Natl Acad Sci U S A 112: 13390-13395 (read)
Dupoiron S, Zischek C, Ligat L, Carbonne J, Boulanger A, Dugé de Bernonville T, Lautier M, Rival P, Arlat M, Jamet E, Lauber E, Albenne C (2015) The N-glycan cluster from Xanthomonas campestris pv. campestris : a toolbox for sequential plant N-glycan processing. J Biol Chem 290: 6022-6036 (read)
Francin-Allami M, Merah K, Albenne C, Rogniaux H, Pavlovic M, Lollier V, Sibout R, Guillon F, Jamet E, Larré C (2015) Cell wall proteomics of Brachypodium distachyon grains : A focus on cell wall remodeling proteins. Proteomics 15: 2296-2306 (read)
Francoz E, Ranocha P, Burlat V, Dunand C (2015) Arabidopsis seed mucilage 1 secretory cells : Regulation and dynamics. Trends Plant Sci 20 : 515-524 (read)
Francoz E, Ranocha P, Nguyen-Kim H, Jamet E, Burlat V, Dunand C (2015) Roles of cell wall peroxidases in plant development. Phytochemistry 112: 15-21 (read)
Lauressergues D, Couzigou JM, San Clemente H, Martinez Y, Dunand, Bécard G, Combier JP (2015) Primary transcripts of microRNAs encode regulatory peptides. Nature 520: 90-93 (read)
Li Q, Yu H, Cao PB, Fawal N, Mathé C, Azar S, Cassan-Wang H, Myburg AA, Grima-Pettenati J, Marque C, Teulières C, Dunand C (2015) Explosive tandem and segmental duplications of multigenic families in Eucalyptus grandis. Genome Biol Evol 7: 1068-1081 (read)
Pont-Lezica RF (2015) Localizing proteins by tissue printing. Methods Mol Biol 1312: 93-104 (read)
Raggi S, Ferrarini A, Delledonne M, Dunand C, Ranocha P, De Lorenzo G, Cervone F, Ferrari S (2015) The Arabidopsis class III peroxidase AtPRX71 negatively regulates growth under physiological conditions and in response to cell wall damage. Plant Physiol 169: 2513-2525 (read)
San Clemente H, Jamet E (2015) WallProtDB, a database resource for plant cell wall proteomics. Plant Methods 11: 2 (read)
Sénéchal F, L’Enfant M, Domon JM, Rosiau E, Crépeau MJ, Surcouf O, Esquivel-Rodriguez J, Marcelo P, Mareck A, Guérineau F, Hyung-Rae K, Mravec J, Bonnin E, Jamet E, Kihara D, Lerouge P, Ralet MC, Pelloux J, Rayon C (2015) Tuning of pectin methylesterification : PECTIN METHYLESTERASE INHIBITOR 7 modulates the processive activity of co-expressed PECTIN MEHYLESTERASE 3 in a pH-dependent manner. J Biol Chem 290: 23320-23335 (read)
Zhu X, Dunand C, Snedden W, Galaud JP (2015) CaM and CMLs emergence in the green lineage. Trends Plant Sci 20: 483-489 (read)
2014
Albenne C, Canut H, Hoffmann L, Jamet E (2014) Plant cell wall proteins : a large body of data, but what about runaways ? Proteomes 2: 224-242 (read)
Calderan-Rodrigues MJ, Jamet E, Calderan Rodrigues Bonassi MB, Guidetti-Gonzalez S, Carmanhanis Begossi A, Vaz Setem L, Franceschini LM, Guimarães Fonseca J, Labate CA (2014) Cell wall proteomics of sugarcane cell suspension cultures. Proteomics 14: 738-749 (read)
Dugé de Bernonville T, Albenne C, Arlat M, Hoffmann L, Lauber E, Jamet E (2014) Xylem sap proteomics. Methods Mol Biol 1072: 391-405 (read)
Fawal N, Li Q, Mathé C, Dunand C (2014) Automatic multigenic family annotation: risks and solutions. Trends Genet 30: 323-325 (read)
Hijazi M, Roujol D, Nguyen-Kim H, del Rocio Cisneros Castillo L, Saland E, Jamet E, Albenne C (2014) Arabinogalactan protein 31 (AGP31), a putative network-forming protein in Arabidopsis thaliana cell walls ? Ann Bot 114: 1087-1097 (read)
Hijazi M, Velasquez MS, Jamet E, Estevez JM, Albenne C (2014) An update on post-translational modifications of hydroxyproline-rich glycoproteins : Towards a model highlighting their contribution to plant cell wall architecture. Front Plant Sci 5: 395 (read)
Lannoo N, Van Damme EJM, Albenne C, Jamet E (2014) Plant Glycobiology – a diverse world of lectins, glycoproteins, glycolipids and glycans. Front Plant Sci 5: 604 (read). Access to e-book.
Myburg AA et al. (2014) The genome of Eucalyptus grandis. Nature 510: 356-362 (read)
Ranocha P, Francoz E, Burlat V, Dunand C (2014) Expression of PRX36, PMEI6 and SBT1.7 is controlled by complex transcription factor regulatory networks for proper seed coat mucilage extrusion. Plant Signal Behav 9: e977734 (read)
Yu H, Soler M, Mila I, San Clemente H, Dunand C, Paiva JAP, Myburg AA, Bouzayen M, Grima-Pettenati J, Cassan-Wang H (2014) Genome-wide characterization and expression profiling of the AUXIN RESPONSE FACTOR (ARF) gene family in Eucalyptus grandis. PLoS One 9: e108906 (read)
2013
Albenne C, Canut H, Jamet E (2013) Plant cell wall proteomics : the leadership of Arabidopsis thaliana. Front Plant Sci 4: 111 (read)
Douché T, San Clemente H, Burlat V, Roujol D, Valot B, Zivy M, Pont-Lezica R, Jamet E (2013) Brachypodium distachyon as a model plant towards improved biofuel crops : Search for secreted proteins involved in biogenesis and disassembly of cell wall polymers. Proteomics 13: 2438-2454 (read)
Fawal N, Li Q, Savelli B, Brette M, Passaia G, Fabre M, Mathé C, Dunand C (2013) PeroxiBase: a database for large-scale evolutionary analysis of peroxidases. Nucleic Acids Res 41(Database issue): D441-444 (read)
Lariguet P, Ranocha P, De Meyer M, Barbier O, Penel C, Dunand C (2013) Identification of a hydrogen peroxide signalling pathway in the control of light-dependent germination in Arabidopsis. Planta 238: 381-395 (read)
Mallèvre F, Roget A, Kervella Y, Ropartz D, Ralet M-C, Canut H, Minon T, Livache T (2013) Microwave heating for the rapid generation of glycosylhydrazides. Bioconjugate Chem 24: 1264-1269 (read)
Simkin AJ, Miettinen K, Claudel P, Burlat V, Guirimand G, Courdavault V, Papon N, Meyer S, Godet S, St-Pierre B, Giglioli-Guivarc’h N, Fischer MJ, Memelink J, Clastre M (2013) Characterization of the plastidial geraniol synthase from Madagascar periwinkle which initiates the monoterpenoid branch of the alkaloid pathway in internal phloem associated parenchyma. Phytochemistry 85: 36-43
2012
Delaux PM, Nanda AK, Mathé C, Sejalon-Delmas N, Dunand C (2012) Molecular and biochemical aspects of plant terrestrialization. Perspect Plant Ecol Evol Syst 14: 49-59 (read)
Delaux PM, Xie X, Timme RE, Puech-Pagès V, Dunand C, Lecompte E, Delwiche CF, Yoneyama K, Bécard G and Séjalon-Delmas N (2012) Origin of strigolactones in the green lineage. New Phytol 195: 857-871 (read)
Fawal N, Savelli B, Dunand C, Mathé C (2012) GECA: a fast tool for Gene Evolution and Conservation Analysis in eukaryotic protein families. Bioinformatics 28: 1398-1399 (read)
Geu-Flores F, Sherden NH, Courdavault V, Burlat V, Glenn WS, Wu C, Nims E, Cui Y, O’Connor SE (2012) An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature 492: 138-142
Guirimand G, Guihur A, Phillips M, Oudin A, Glévarec G, Mahroug S, Melin C, Papon N, Clastre M, Giglioli-Guivarc’h N, St-Pierre B, Rodríguez-Concepción M, Burlat V, Courdavault V (2012) Triple subcellular targeting of isopentenyl diphosphate isomerases encoded by a single gene. Plant Signal Behav 7: 1495-1497
Guirimand G, Guihur A, Phillips MA, Oudin A, Glévarec G, Melin C, Papon N, Clastre M, St-Pierre B, Rodríguez-Concepción M, Burlat V, Courdavault V (2012) A single gene encodes isopentenyl diphosphate isomerase isoforms targeted to plastids, mitochondria and peroxisomes in Catharanthus roseus. Plant Mol Biol 79: 443-459
Guirimand G, Simkin AJ, Papon N, Besseau S, Burlat V, St-Pierre B, Giglioli-Guivarc’h N, Clastre M, Courdavault V (2012) Cycloheximide as a tool to investigate protein import in peroxisomes : a case study of the subcellular localization of isoprenoid biosynthetic enzymes. J Plant Physiol 169: 825-829
Hijazi M, Durand J, Pichereaux C, Pont F, Jamet E, Albenne C (2012) Characterization of the ArabinoGalactan Protein 31 (AGP31) of Arabidopsis thaliana : new advances on the Hyp-O-glycosylation of the Pro-rich domain. J Biol Chem 287: 9623-9632 (read)
Marino D, Dunand C, Puppo A, Pauly N (2012) A burst of plant NADPH oxidases. Trends Plant Sci 17: 9-15 (read)
Olson A, Aerts,A, Asiegbu F, Belbahri L, Bouzid O, Broberg A, Canbäck B, Coutinho PM, Cullen D, Dalman K, Deflorio G, van Diepen L, Dunand C, Duplessis S., Durling M., Gonthier P, Grimwood J, Gunnar Fossdal C, Hansson, D., Henrissat, B., Hietala, A., Himmelstrand, K., Hoffmeister D, Hogberg N, James T, Karlsson J, Kohler A, Kües U, Lee Y, Lin Y-C, Lind M, Lindquist E, Lombard V, Lucas S, Lunden K, Morin E, Murat C, Park R, Raffaello T, Rouzé P, Salamov A, Schmutz J, Solheim H, Stahlberg J, Velez M, de Vries R, Wiebenga A, Woodward S, Yakovlev I, Garbelotto M, Martin F, Grigoriev I, Stenlid J (2012) Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytol 194: 1001-1013 (read)
Vázquez-Lobo A, Roujol D, Zuñiga-Sánchez E, Albenne C, Piñero D, Gamboa de Buen A, Jamet E (2012) The highly conserved spermatophyte cell wall DUF642 protein family : phylogeny and first evidence of interaction with cell wall polysaccharides in vitro. Mol Phylogenet Evol 63: 510-520 (read)
2011
Bouwmeester K, de Sain M, Weide R, Gouget A, Klamer S, Canut H, Govers F (2011) The lectin receptor kinase LecRK-I.9 is a novel Phytophthora resistance component and a host target for a RXLR effector. PLoS Pathog 7: e1001327 (read)
Dunand C, Mathé C, Lazzarotto F, Margis R, Margis-Pinheiro M (2011) Ascorbate peroxidase-related (APx-R) is not a duplicable gene. Plant Signal Behav 6: 1908-1913 (read)
Guirimand G, Guihur A, Ginis O, Poutrain P, Héricourt F, Oudin A, Lanoue A, St-Pierre B, Burlat V, Courdavault V (2011) The subcellular organization of strictosidine biosynthesis in Catharanthus roseus epidermis highlights several trans-tonoplast translocations of intermediate metabolites. FEBS J 278: 749-63
Guirimand G, Guihur A, Poutrain P, Héricourt F, Mahroug S, St-Pierre B, Burlat V, Courdavault V (2011) Spatial organization of the vindoline biosynthetic pathway in Catharanthus roseus. J Plant Physiol 6: 549-557
Guénin S, Mareck A, Rayon C, Lamour R, Assoumou, Ndong Y, Domon JM, Sénéchal F, Fourneta F, Jamet E , Canut H, Percoco G, Mouille G, Rolland A, Rustérucci C, Guérineau F, Van Wuytswinkel O, Gillet F, Driouich A, Lerouge P, Gutierrez L, Pelloux J (2011) Identification of PME3 as a basic pectin methylesterase isoform involved in adventitious rooting in Arabidopsis. New Phytol 192: 114-126 (read)
Jamet E (2011) Obituary : Rafael Pont-Lezica. Front Plant Sci 2: 86 (read)
Lazzarotto F, Teixeira FK, Barcelos Rosa S, Dunand C, Lemelle Fernandes C, de Vasconcelos Fontenele A, Silveira JAG, Verli H, Margis R, Margis-Pinheiro M (2011) APX-R is a new heme-containing protein functionally associated to APx but evolutionarily divergent. New Phytol 191: 234-250 (read)
Ligat L, Lauber E, Albenne C, San Clemente H, Valot B, Zivy M, Pont-Lezica R, Arlat M, Jamet E (2011) Analysis of the xylem sap proteome of Brassica oleracea reveals a high content in secreted proteins. Proteomics 11: 1798-1813 (read)
Poutrain P, Guirimand G, Mahroug S, Burlat V, Melin C, Ginis O, Oudin A, Giglioli-Guivarc’h N, Pichon O, Courdavault V (2011) Molecular cloning and characterisation of two calmodulin isoforms of the Madagascar periwinkle Catharanthus roseus. Plant Biol 13: 36-41
Zhang Y, Giboulot A, Zivy M, Valot B, Jamet E, Albenne C (2011) Combining various strategies to increase the coverage of the plant cell wall glycoproteome. Phytochemistry 72: 1109-1123 (read)
2010
Cosio C, Dunand C (2010) Transcriptome analysis of various flower and silique development stages indicates a set of class III peroxidase genes potentially involved in pod shattering in Arabidopsis thaliana. BMC Genomics 11: 528 (read)
Guirimand G, Courdavault V, Lanoue A, Mahroug S, Guihur A, Blanc N, Giglioli-Guivarc’h N, St-Pierre B, Burlat V (2010) Strictosidine activation in Apocynaceae : towards a “nuclear time bomb”? BMC Plant Biol 10 : 20
Guirimand G, Courdavault V, St-Pierre B, Burlat V (2010) Biosynthesis and regulation of alkaloids. In E Pua, M Davey, eds, Plant developmental biology – Biotechnological perspectives, vol 2. Springer-Verlag, Berlin Heidelberg, pp 139-160
Jamet E, Pont-Lezica R (2010) The puzzle of protein location in plant proteomics. In G Rancourt, ed, Proteomics : Methods, Applications and Limitations. Nova Science Publishers, Inc., Hauppauge, NY, USA (read)
Lanoue A, Burlat V, Henkes GJ, Koch I, Schurr U, Rose USR (2010) De novo biosynthesis of defense root exudates in response to Fusarium attack in barley. New Phytol 185 : 577-588 (read)
Lanoue A, Burlat V, Schurr U, Rose US (2010) Induced root-secreted phenolic compounds as a belowground plant defense. Plant Signal Behav 5: 1037-1038
Mathé C, Barre A, Jourda C, Dunand C (2010) Evolution and expression of class III peroxidases. Arch Biochem Biophys 500: 58-65 (read)
Nanda AK, Andrio E, Marino D, Pauly N, Dunand C (2010) Reactive oxygen species during plant-microorganism early interactions. J Integr Plant Biol 52: 195-204 (read)
Pont-Lezica R, Minic Z, San Clemente H, Roujol D, Jamet E (2010) Plant cell wall functional genomics : Novelties from proteomics. In M Osborne, ed, Advances in Genetics Research, vol 1. Nova Science Publishers, Inc, Hauppauge, NY, USA, pp 265-281 (read)
2009
Albenne C, Canut H, Boudart G, Zhang Y, San Clemente H, Pont-Lezica R, Jamet E (2009) Plant cell wall proteomics : mass spectrometry data, a trove for research on protein structure/function relationships. Mol Plant 2: 977-989 (read)
Cosio C, Dunand C (2009) Specific functions of individual class III peroxidase genes. J Exp Bot 60: 391-408 (read)
Cosio C, Vuillemin L, De Meyer M, Kevers C, Penel C, Dunand C (2009) An anionic class III peroxidase from zucchini may regulate hypocotyl elongation through its auxin oxidase activity. Planta 229: 823-836 (read)
Courdavault V, Burlat V, St-Pierre B, Giglioli-Guivarc’h N (2009) Proteins prenylated by type I protein geranylgeranyltransferase act positively on the jasmonate signalling pathway triggering the biosynthesis of monoterpene indole alkaloids in Catharanthus roseus. Plant Cell Rep 28: 83-93
Guirimand G, Burlat V, Oudin A, Lanoue A, St-Pierre B, Courdavault V (2009) Optimization of the transient transformation of Catharanthus roseus cells by particle bombardment and its application to the subcellular localization of hydroxymethylbutenyl 4-diphosphate synthase and geraniol 10-hydroxylase. Plant Cell Rep 28: 1215-1234
Jamet E, Roujol D, San Clemente H, Irshad M, Soubigou-Taconnat L, Renou J, Pont-Lezica R (2009) Cell wall biogenesis of Arabidopsis thaliana elongating cells : transcriptomics complements proteomics. BMC Genomics 10: 505 (read)
Koua D, Cerutti L, Falquet L, Sigrist CJA, Theiler G, Hulo N, Dunand C (2009) PeroxiBase: a database with new tools for peroxidase family classification. Nucleic Acids Res 37: D261-D266 (read)
Minic Z, Jamet E, San Clemente H, Pelletier S, Renou J, Rihouey C, Okinyo D, Proux C, Lerouge P, Jouanin L (2009) Transcriptomic analysis of Arabidopsis developing stems : a close-up on cell wall genes. BMC Plant Biol 9: 17 (read)
Oliva M, Theiler G, Zamocky M, Koua D, Margis-Pinheiro M, Passardi F, Dunand C (2009) PeroxiBase: a powerful tool to collect and analyse peroxidase sequences from Viridiplantae. J Exp Bot 60: 453-459 (read)
Pont-Lezica RF (2009) Localizing proteins by tissue printing. Methods Mol Biol 536: 75-88 (read)
San Clemente H, Pont-Lezica R, Jamet E (2009) Bioinformatics as a tool for assessing the quality of sub-cellular proteomic strategies and inferring functions of proteins : plant cell wall proteomics as a test case. Bioinform Biol Insights 3: 15-28 (read)
2008
Foissac S, Gouzy J, Rombauts S, Mathé C, Amselem J, Sterck L, Van de Peer Y, Rouzé P, Schiex T (2008) Genome annotation in plants and fungi : EuGene as a model platform. Curr Bioinform 3: 87-97 (read)
Irshad M, Canut H, Borderies G, Pont-Lezica R, Jamet E (2008) A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana : confirmed actors and newcomers. BMC Plant Biol 8: 94 (read)
Jamet E, Albenne C, Boudart G, Irshad M, Canut H, Pont-Lezica R (2008) Recent advances in plant cell wall proteomics. Proteomics 8: 893-908 (read)
Jamet E, Boudart G, Borderies G, Charmont S, Lafitte C, Rossignol M, Canut H, Pont-Lezica R (2008) Isolation of plant cell wall proteins. Methods Mol Biol 425: 187-201 (read)
Jamet E, Canut H, Albenne C, Boudart G, Pont-Lezica R (2008) Cell wall. In G Agrawal, R Rakwal, eds, Plant Proteomics : Technologies, Strategies, and Applications. Wiley, USA, pp 293-307 (read)
Jouili H, Bouazizi H, Rossignol M, Borderies G, Jamet E, El Ferjani E (2008) Partial purification and characterization of a copper-induced anionic peroxidase of sunflower roots. Plant Physiol Biochem 46: 760-767 (read)
Margis R, Dunand C, Teixeira FK, Margis-Pinheiro M (2008) Glutathione peroxidase family – an evolutionary overview. FEBS J 275: 3959-3970 (read)
Zamocky M, Jakopitsch C, Furtmuller PG, Dunand C, Obinger C (2008) The peroxidase-cyclooxygenase superfamily: reconstructed evolution of critical enzymes of the innate immune system. Proteins 72: 589-605 (read)
2007
Boudart G, Minic Z, Albenne C, Canut H, Jamet E, Pont-Lezica R (2007) Cell wall proteome. In S Samaj, J Thelen, eds, Plant proteomics. Springer, Berlin, pp 169-185 (read)
Minic Z, Jamet E, Négroni L, der Garabedian AP, Zivy M, Jouanin L (2007) A sub-proteome of Arabidopsis thaliana mature stems trapped on Concanavalin A is enriched in cell wall glycoside hydrolases. J Exp Bot 58: 2503-2512 (read)
2006
Feiz L, Irshad M, Pont-Lezica R, Canut H, Jamet E (2006) Evaluation of cell wall preparations for proteomics : a new procedure for purifying cell walls from Arabidopsis hypocotyls. Plant Methods 2 : 10 (read)
Gouget A, Senchou V, Govers F, Sanson A, Barre A, Rougé P, Pont-Lezica R, Canut H (2006) Lectin receptor kinases participate in protein-protein interactions to mediate plasma membrane-cell wall adhesions in Arabidopsis. Plant Physiol 140 : 81-90 (read)
Jamet E, Canut H, Boudart G, Pont-Lezica R (2006) Cell wall proteins : a new insight through proteomics. Trends Plant Sci 11 : 33-39 (read)
2005
Aubourg S, Brunaud V, Bruyere C, Cock M, Cooke R, Cottet A, Couloux A, Dehais P, Deleage G, Duclert A, Echeverria M, Eschbach A, Falconet D, Filippi G, Gaspin C, Geourjon C, Grienenberger J-M, Houlné G, Jamet E, Lechauve F, Leleu O, Leroy P, Mache R, Meyer C, Nedjari H, Negrutiu I, Orsini V, Peyretaillade E, Pommier C, Raes J, Risler J-L, Rivière S, Rombault S, Rouzé P, Schneider M, Schwob P, Small I, Soumayet-Kampetanga G, Stankovski D, Toffano C, Tognolli M, Caboche M, Lecharny A (2005) GENEFARM, structural and functional annotation of Arabidopsis gene and protein families by a network of experts. Nucleic Acids Res 33 : D641-D646. (read)
Boudart G, Jamet E, Rossignol M, Lafitte C, Borderies G, Jauneau A, Esquerré-Tugayé M, Pont-Lezica R (2005) Cell wall proteins in apoplastic fluids of Arabidopsis thaliana rosettes : identification by mass spectrometry and bioinformatics. Proteomics 5 : 212-221. (read)
Charmont S, Jamet E, Pont-Lezica R, Canut H (2005) Proteomic analysis of secreted proteins from Arabidopsis thaliana seedlings : improved recovery following removal of phenolic compounds. Phytochemistry 66 : 453-461. (read)
Masclaux F, Galaud J, Pont-Lezica R (2005) The riddle of the plant vacuolar sorting receptors. Protoplasma 226 : 103-108. (read)
Masclaux F, Pont-Lezica R, Galaud J (2005) Relationship between allelic state of T-DNA and DNA methylation of chromosomal integration region in transformed Arabidopsis thaliana plants. Plant Mol Biol 58 : 295-303. (read)
Schantz M, Jamet E, Guitton A, Schantz R, Houlné G (2005) Functional analysis of the bell pepper knolle gene (cakn) promoter region in tobacco plants and in synchronized BY2 cells. Plant Sci 169 : 155-163. (read)
2004
Guzzardi P, Genot G, Jamet E (2004) The Nicotiana sylvestris extensin gene, Ext 1.2A, is expressed in the root transition zone and upon wounding. Biochim Biophys Acta 1680 : 83-92. (read)
Jamet E (2004) Bioinformatics as a critical prerequisite to transcriptome and proteome studies. J Exp Bot 55 : 1977-1979. (read)
Masclaux F, Charpenteau M, Takahashi T, Pont-Lezica R, Galaud J (2004) Gene silencing using a heat-inducible RNAi system in Arabidopsis. Biochem Biophys Res Commun 321 : 364-369. (read)
Senchou V, Weide R, Carrasco A, Bouyssou H, Pont-Lezica R, Govers F, Canut H (2004) High affinity recognition of a Phytophthora protein by Arabidopsis via an RGD motif. Cell Mol Life Sci 61 : 502-509. (read)
2003
Borderies G, Jamet E, Lafitte C, Rossignol M, Jauneau A, Boudart G, Monsarrat B, Esquerré-Tugayé M, Boudet A, Pont-Lezica R (2003) Proteomics of loosely bound cell wall proteins of Arabidopsis thaliana cell suspension cultures : a critical analysis. Electrophoresis 24 : 3421-3432. (read)
Laval V, Masclaux F, Serin A, Carrière M, Roldan C, Devic M, Pont-Lezica R, Galaud J (2003) Seed germination is blocked in Arabidopsis putative vacuolar sorting receptor (atbp80) antisense transformants. J Exp Bot 54 : 213-221 (read)
Salva I, Jamet E (2003) The tobacco Ext 1.4 extensin gene family is not regulated in cells proliferating under hormone control Plant Physiol Biochem 41 : 363-367 (read)
Theses
Antoine FIRMIN (2019-2023) Institut National Polytechnique de Toulouse. La mixotrophie chez les plantes aquatiques : étude expérimentale d’un métabolisme méconnu.
Bastien DAUPHIN (2019-2022). Université Paul Sabatier-Toulouse 3. Dynamique de microdomaines pariétaux pendant le développement des cellules sécrétrices du mucilage.
Hasan KOLKAS (2019-2022). Université Paul Sabatier-Toulouse 3. Les protéines pariétales à domaine PAC dans la lignée verte : Rôle dans l’architecture des parois végétales.
Ali ELJEBBAWI (2018-2021). Université Paul Sabatier-Toulouse 3. Arabidopsis thaliana : un modèle pour étudier les adaptations thermique et saline des plantes dans les Pyrénées. (read)
Sébastien VIUDES (2018-2021). Université Paul Sabatier-Toulouse 3. Variabilité naturelle des mucilages des graines de Brassicaceae : étude comparative inter et intra espèces. (read)
Maxime BAFOIL (2018-2020). Université Paul Sabatier-Toulouse 3. Stimulation de la germination des graines et de leur croissance par plasmas froids à la pression atmosphérique. (read)
Duchesse Lacours MBADINGA MBADINGA (2017-2020). Université Paul Sabatier-Toulouse 3. Evolution de la cutine chez les plantes et son rôle durant la terrestrialisation.
Kevin BELLANDE (2015-2018). Université Paul Sabatier-Toulouse 3. Etude fonctionnelle d’un récepteur lectine kinase, LecRK-I.9 : un contrôle de la dynamique des parois chez Arabidopsis thaliana.
Harold DURUFLE (2014-2017). Université Paul Sabatier-Toulouse 3. Production et traitement de données « omics » hétérogènes en vue de l’étude de la plasticité de la paroi chez des écotypes pyrénéens de la plante modèle Arabidopsis thaliana. (read)
Prix de thèse Henri Gaussen de l’Académie des Sciences Inscriptions et Belles lettres de Toulouse (2018)
Adélaïde JACQ (2013-2016). Université Paul Sabatier-Toulouse 3. Caractérisation fonctionnelle d’AtLTP2, une protéine de transfert de lipides impliquée dans le contrôle de l’intégrité de la cuticule chez Arabidopsis thaliana. (read)
Edith FRANCOZ (2012-2015). Université Paul Sabatier, Toulouse 3. Rôle de la famille multigénique des peroxydases pariétales de classe III dans le développement et la dynamique des parois cellulaires végétales. (read)
Prix de thèse Henri Gaussen de l’Académie des Sciences Inscriptions et Belles lettres de Toulouse (2016) (read)
Huan NGUYEN-KIM (2011-2015). Université Paul Sabatier-Toulouse 3. Recherche de la fonction de protéines riches en hydroxyproline dans les parois végétales. (read)
Qiang LI (2011-2014). Université Paul Sabatier-Toulouse 3. Global evolutionary analysis of CIII peroxidases in the green lineage. (read)
Nizar FAWAL (2010-2013). Université Paul Sabatier-Toulouse 3. Automatic annotation of multigene families, the case of peroxidases.(read)
May HIJAZI (2008-2011). Université Paul Sabatier-Toulouse 3. Caractérisation structurale et fonctionnelle d’AGP31, une glycoprotéine atypique de la paroi chez Arabidopsis thaliana. (read)
Muhammad IRSHAD (2005-2008). Université Paul Sabatier-Toulouse 3. Dynamique des protéines pariétales au cours de l’élongation cellulaire dans des hypocotyles étiolés d’Arabidopsis thaliana : approches protéomique et transcriptomique. (read)
Anne GOUGET (2003-2006). Université Paul Sabatier-Toulouse 3. Etude fonctionnelle d’un récepteur lectine kinase (LecRK79), potentiel partenaire dans les contacts paroi-plasmalemme chez Arabidopsis thaliana. (read)
lrsv.gestion@univ-tlse3.fr
Phone
Standard : 05.34.32.38.01
Fax : 05.34.32.38.02
Find us
24, chemin de Borde-Rouge.
31320, Auzeville-Tolosane. FRANCE