Our Team
Fabien MOUNET
Maître de Conférence UPS - Team manager
fabien.mounetATuniv-tlse3DOTfr

Chantal TEULIERES
Professeur UPS
marie-chantal.teuliereATuniv-tlse3DOTfr

Hua WANG-CASSAN
Maître de Conférence UPS
hua.cassanATuniv-tlse3DOTfr

Nathalie LADOUCE
Assistante Ingénieur CNRS
nathalie.ladouceATuniv-tlse3DOTfr

Annabelle DUPAS
Assistante Ingenieur UPS
annabelle.dupasATuniv-tlse3DOTfr
Former members
Ines HADJ BACHIR (Doctorante 2019-2022)
Jacqueline GRIMA-PETTENATI (Chercheuse CNRS – Directrice d’équipe)
Maylis GALLOIS (post-doctorante)
Ying DAI (Doctorante)
Christiane MARQUE (Maître de conférence UPS)
Camille RIBEYRE ( CDD- Assistante Ingénieure)
Raphaël PLOYET (doctorant 2013-2016)
Isabelle TRUCHET ( Maître de Conférence UPS )
Jorge LEPIKSON-NETO (post-doc, 2014-2015 )
Marçal SOLER (post-doc, 2010-2015 )
Anna PLASENCIA (doctorante SEVAB, 2012-2015)
Chien NGUYEN (doctorante SEVAB, 2012-2015)
Hong YU (doctorante SEVAB, 2010-2014)
Eveline TAVARES (doctorante, 2014)
Bang CAO (doctorant SEVAB, 2010-2013)
Annette NASSUTH (Professeur invité, Guelph university, Canada, 2012-2013)
Audrey COURTIAL (doctorante 2009-2012)
Sahar AZAR (doctorante 2009-2012)
Mohammed Najib SAIDI (post-doc 2011-2012)
Nadia GOUE (post-doc 2010-2012)
Eduardo CAMARGO (doctorante Brésilien 2010-2011)
Malha AMGOUD (doctorante Algérienne 2010-2011)
Nicolas AMELOT (doctorant 2006-2010)
Marie NAVARRO (doctorante 2005-2009)
Gilles MARQUE (doctorant 2004-2008)
Sylvain LEGAY (doctorant 2003-2008)
Pierre SIVADON (maître de conférence Université Pau)
Research Topics
Background and objectives
Wood, the most abundant lignocellulosic biomass on earth, is widely used for lumber and paper manufacture and increasingly exploited as an environmentally cost-effective, renewable source of energy. The major technological barrier to the efficient use of lignocellulosic biomass for biofuels is the secondary cell walls’ (SCW) recalcitrance to degradation.
The main goal of the “Eucalyptus functional genomics” group is to understand the regulation of wood formation during development and in response to environmental cues. While improving our basic knowledge of this complex process, we aim at identifying regulatory genes, which control wood properties in order to select/design trees more adapted to specific wood end-uses.
Our target plant is Eucalyptus, the first planted forest tree in the world [20M ha] and the second whose genome has been sequenced [Myburg et al, 2014].
- New regulators of wood biosynthesis
- Regulatory networks underlying wood formation under nutritional and abiotic stress
- Croos talk between cold tolerance, biomass production and wood formation
- Regulating the regulators: Insights into the SCW regulating Interactome
For instance, the comparative phylogenetic analysis of the R2R3 MYB stressed out some subgroups that were more expanded in woody plants than in herbaceous plants, and strikingly new subgroups that seem to contain only members from woody plants, being absent in Arabidopsis and rice. Remarkably, 94% of the tandem duplicated R2R3-MYB genes belong to those woody-expanded and woody-preferential subgroups. All members of three woody-preferential subgroups are preferentially expressed in secondary cambium. Cambial activity is responsible for secondary-growth, one major feature of woody perennial plants.

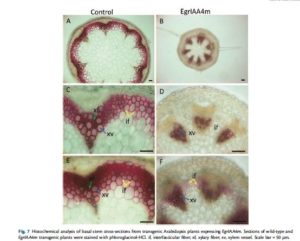
A major outcome of this work was to highlight promising candidate genes possibly involved in the control of wood formation during development and in responses to stresses. We chose some of these candidate genes according to their expression patterns and novelty for functional characterization and transformed Arabidopsis and poplar lines with overexpressing and/or dominant repressor constructs. For instance, the ectopic expression of stabilized Eucalyptus Aux/IAA gene EgIAA4 in Arabidopsis impressively reduced primary and lateral growth lead to drawf plant with very thin stem, especially greatly inhibited interfascular fiber development (Figure below, Yu et al 2015). The overexpression of other two Eucalyptus Aux/IAA genes in Arabidopsis also greatly reduce fiber cell development.

We also implemented E. grandis hairy root systems suitable for medium throughput functional studies of genes involved in secondary cell wall biosynthesis. As a proof-of-concept, we down-regulated the Cinnamoyl-CoA Reductase1 (EgCCR1) gene [Plasencia et al, 2015] and analyzed roots using histochemical, transcriptomic and biochemical approaches.
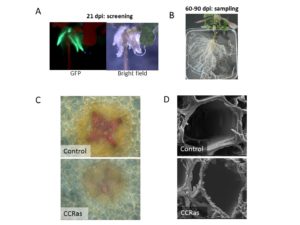
Fig . Development and validation of a fast system for functional characterization of candidate genes in a homolog system of Eucalyptus using hairy roots.
A. Eucalyptus plants transformed with A. rhizogenes harbouring a plasmid with 35S:GFP were detected under fluorescence 21 days after infection. B. Composite plants resulting of the A. rhizogenes transformation were hardened in pots with inert substrate and are ready to be harvested after 60-90 days after infection. C. Suitability of this system was assessed by transforming Eucalyptus with the antisense construct of the CCR gene, showing hairy roots section stained with phloroglucinol reagent with thinner xylem cell walls and lower lignin content. D. Deeper analysis using Scanning Electron Microscopy demonstrated thinner xylem cell walls with a phenotype resembling to irregular xylem, as previously observed (Piquemal et al. 1998).
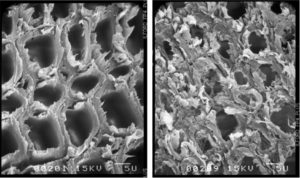

Fig SEM images of Eucalyptus xylem SCW under limiting (left) and luxuriant (right) nitrogen fertilization conditions [Camargo et al, 2014]
Fig SEM images of Eucalyptus xylem SCW under [Ployet et al, 2017]
We develop system biology approaches aiming to integrate transcriptomic, metabolomic and epigenomic data in order to highlight regulatory pathways involved in cold or drought stress.



Publications
2022
- Karannagoda, N.; Spokevicius, A.; Hussey, S.; Cassan-Wang, H.; Grima-Pettenati, J.; Bossinger, G. (2022) Eucalyptus grandis AUX/INDOLE-3-ACETIC ACID 13 (EgrIAA13) is a novel transcriptional regulator of xylogenesis. Plant Mol Biol 2022, doi:10.1007/s11103-022-01255-y.
- H Yu, M Li, Z Zhu, A Wu, F Mounet, E Pesquet, J Grima-Pettenati, H Cassan-Wang, (2022) Overexpression of EgrIAA20 from Eucalyptus grandis, a Non-Canonical Aux/IAA Gene, Specifically Decouples Lignification of the Different Cell-Types in Arabidopsis Secondary Xylem. Int. J. Mol. Sci. 2022, 23, 5068.https://doi.org/10.3390/ijms23095068
2021
- Dumond L, Lam PY, Van Erven G, Kabel M, Mounet F, Grima-Pettenati J, Tobimatsu Y, Hernandez-Raquet G (2021) Termite Gut Microbiota Contribution to Wheat Straw Delignification in Anaerobic Bioreactors. ACS Sustainable Chem. Eng. 9, 5, 2191–2202
2020
- Dai Y, Hu G, Dupas A, Medina L, Blandels N, Clemente HS, Ladouce N, Badawi M, Hernandez-Raquet G, Mounet F, Grima-Pettenati J, Cassan-Wang H (2020) Implementing the CRISPR/Cas9 Technology in Eucalyptus Hairy Roots Using Wood-Related Genes. Int J Mol Sci 21
- Cao PB, Ployet R, Nguyen C, Dupas A, Ladouce N, Martinez Y, Grima-Pettenati J, Marque C, Mounet F, Teulieres C (2020) Wood Architecture and Composition Are Deeply Remodeled in Frost Sensitive Eucalyptus Overexpressing CBF/DREB1 Transcription Factors. Int J Mol Sci 21
2019
- Gomes C, Dupas A, Pagano A, Grima-Pettenati J, Paiva JAP (2019) Hairy Root Transformation: A Useful Tool to Explore Gene Function and Expression in Salix spp. Recalcitrant to Transformation. Front Plant Sci 10: 1427
- Ployet R, Venezio-Labate MT, Regiani Catalidi T, Christina M, Morel M, San Clemente H, Denis M, Favreau B, Tomazello Filho M, Laclau JP, Labate CA, Chaix G, Grima-Pettenati J and F Mounet (2019). A system biology viw of wood formation in Eucalyptus grandis trees submitted to different potassium and water regimes. New Phytol .
- Camargo E, Ployet R, Cassan-Wang H, Mounet F, Grima-Pettenati J. 2019. Digging in wood: New insights in the regulation of wood formation in tree species. Advances in Botanical Research 89: 201-233.
- Hussey SG, Grima-Pettenati J, Myburg AA, Mizrachi E, Brady SM, Yoshikuni Y, Deutsch S. (2019) A Standardized Synthetic Eucalyptus Transcription Factor and Promoter Panel for Re-engineering Secondary Cell Wall Regulation in Biomass and Bioenergy Crops. Acs Synthetic Biology 8(2): 463-465.
- Lin Z, Long JM, Yin Q, Wang B, Li HL, Luo JZ, Wang HC, Wu AM.(2019). Identification of novel lncRNAs in Eucalyptus grandis. Industrial Crops and Products 129: 309-317.
- Tavares EQP, De Souza AP, Romim GH, Grandis A, Plasencia A, Gaiarsa JW, Grima-Pettenati J, de Setta N, Van Sluys MA, Buckeridge MS.(2019). The control of endopolygalacturonase expression by the sugarcane RAV transcription factor during aerenchyma formation. Journal of Experimental Botany 70(2): 497-506.
- Favreau B, Denis M, Ployet R, Mounet F, Da Silva HP, Franceschini L, Laclau JP, Labate C, Carrer H (2019) Distinct leaf transcriptomic response of water deficient Eucalyptus grandis submitted to potassium and sodium fertilization. PLoS One 14(6): e0218528
2018
- Plomion C, Aury JM, Amselem J, Leroy T, Murat F, Duplessis S, Faye S, Francillonne N, Labadie K, Le Provost G, Lesur I, Bartholome J, Faivre-Rampant P, Kohler A, Leple JC, Chantret N, Chen J, Dievart A, Alaeitabar T, Barbe V, Belser C, Berges H, Bodenes C, Bogeat-Triboulot MB, Bouffaud ML, Brachi B, Chancerel E, Cohen D, Couloux A, Da Silva C, Dossat C, Ehrenmann F, Gaspin C, Grima-Pettenati J, Guichoux E, Hecker A, Herrmann S, Hugueney P, Hummel I, Klopp C, Lalanne C, Lascoux M, Lasserre E, Lemainque A, Desprez-Loustau ML, Luyten I, Madoui MA, Mangenot S, Marchal C, Maumus F, Mercier J, Michotey C, Panaud O, Picault N, Rouhier N, Rue O, Rustenholz C, Salin F, Soler M, Tarkka M, Velt A, Zanne AE, Martin F, Wincker P, Quesneville H, Kremer A, Salse J. 2018. Oak genome reveals facets of long lifespan. Nat Plants 4(7): 440-452
- Xu C, Shen Y, He F, Fu X, Yu H, Lu W, Li Y, Li C, Fan D, Wang HC, Luo K. (2018). Auxin-mediated Aux/IAA-ARF-HB signaling cascade regulates secondary xylem development in Populus. New Phytol.
2017
- Ployet R, Soler M, Carocha V, Ladouce N, Alves A, Rodrigues JC, Harvengt L, Marque C, Teulières C, Grima-Pettenati J, Mounet F (2017) Long cold exposure induces transcriptional and biochemical remodelling of xylem secondary cell wall in Eucalyptus. Tree Physiol: 1-14
- Nguyen HC, Cao PB, San Clemente H, Ployet R, Mounet F, Ladouce N, Harvengt L, Marque C, Teulieres C (2017) Special trends in CBF and DREB2 groups in Eucalyptus gunnii vs Eucalyptus grandis suggest that CBF are master players in the trade-off between growth and stress resistance. Physiol Plant 159: 445-467
2016
- Barrière Y, Courtial A, Chateigner-Boutin AL, Denoue D, Grima-Pettenati J (2016) Breeding maize for silage and biofuel production, an illustration of a step forward with the genome sequence. Plant Science 242: 310-329
- Plasencia A, Soler M, Dupas A, Ladouce N, Silva-Martins G, Martinez Y, Lapierre C, Franche C, Truchet I, Grima-Pettenati J (2016) Eucalyptus hairy roots, a fast, efficient and versatile tool to explore function and expression of genes involved in wood formation. Plant Biotechnology Journal 14: 1381-1393
- Sixto H, Gonzalez-Gonzalez BD, Molina-Rueda JJ, Garrido-Aranda A, Sanchez MM, Lopez G, Gallardo F, Canellas I, Mounet F, Grima-Pettenati J, Canton F (2016) Eucalyptus spp. and Populus spp. coping with salinity stress: an approach on growth, physiological and molecular features in the context of short rotation coppice (SRC). Trees-Structure and Function 30: 1873-1891
- Soler M, Plasencia A, Larbat R, Pouzet C, Jauneau A, Rivas S, Pesquet E, Lapierre C, Truchet I, Grima-Pettenati J (2017) The Eucalyptus linker histone variant EgH1.3 cooperates with the transcription factor EgMYB1 to control lignin biosynthesis during wood formation. New Phytologist 213: 287-299
- Soler M, Plasencia A, Lepikson-Neto J, Camargo ELO, Dupas A, Ladouce N, Pesquet E, Mounet F, Larbat R, Grima-Pettenati J (2016) The Woody-Preferential Gene EgMYB88 Regulates the Biosynthesis of Phenylpropanoid-Derived Compounds in Wood. Frontiers in Plant Science 7
2015
Barriere, Y., A. Courtial, M. Soler and J. Grima-Pettenati (2015). “Toward the identification of genes underlying maize QTLs for lignin content, focusing on colocalizations with lignin biosynthetic genes and their regulatory MYB and NAC transcription factors.” Molecular Breeding 35(3).
Cao, P. B., S. Azar, H. SanClemente, F. Mounet, C. Dunand, G. Marque, C. Marque and C. Teulieres (2015). “Genome-wide analysis of the AP2/ERF family in Eucalyptus grandis : an intriguing over-representation of stress-responsive DREB1/CBF genes.” PLoS One 10(4) : e0121041.
Carocha, V., M. Soler, C. Hefer, H. Cassan-Wang, P. Fevereiro, A. A. Myburg, J. A. Paiva and J. Grima-Pettenati (2015). “Genome-wide analysis of the lignin toolbox of Eucalyptus grandis.” New Phytol 206(4) : 1297-1313.
Hussey, S. G., M. N. Saidi, C. A. Hefer, A. A. Myburg and J. Grima-Pettenati (2015). “Structural, evolutionary and functional analysis of the NAC domain protein family in Eucalyptus.” New Phytol 206(4) : 1337-1350.
Li, Q., H. Yu, P. B. Cao, N. Fawal, C. Mathe, S. Azar, H. Cassan-Wang, A. A. Myburg, J. Grima-Pettenati, C. Marque, C. Teulieres and C. Dunand (2015). “Explosive Tandem and Segmental Duplications of Multigenic Families in Eucalyptus grandis.” Genome Biol Evol 7(4) : 1068-1081.
Soler, M., E. L. Camargo, V. Carocha, H. Cassan-Wang, H. San Clemente, B. Savelli, C. A. Hefer, J. A. Paiva, A. A. Myburg and J. Grima-Pettenati (2015). “The Eucalyptus grandis R2R3-MYB transcription factor family : evidence for woody growth-related evolution and function.” New Phytol 206(4) : 1364-1377.
Yu, H., M. Soler, H. San Clemente, I. Mila, J. A. Paiva, A. A. Myburg, M. Bouzayen, J. Grima-Pettenati and H. Cassan-Wang (2015). “Comprehensive Genome-Wide Analysis of the Aux/IAA Gene Family in Eucalyptus : Evidence for the Role of EgrIAA4 in Wood Formation.” Plant Cell Physiol 56(4) : 700-714.
2014
Yu, H., M. Soler, I. Mila, H. San Clemente, B. Savelli, C. Dunand, J. A. Paiva, A. A. Myburg, M. Bouzayen, J. Grima-Pettenati and H. Cassan-Wang (2014). “Genome-wide characterization and expression profiling of the AUXIN RESPONSE FACTOR (ARF) gene family in Eucalyptus grandis.” PLoS One 9(9) : e108906.
Myburg A, Grattapaglia D, Tuskan G, Hellsten U, Hayes R, Grimwood J, Jenkins J, Lindquist E, Tice H, Bauer D, Goodstein D, Dubchak I, Poliakov A, Mizrachi E, Kullan A, Van Jaarsveld I, Hussey S, Pinard D, Silva-Junior O, Togawa R, Pappas M, Faria D, Sansaloni C, Petroli C, X Yang, P Ranjan, T Tschaplinski, C Ye, Ting Li, L Sterck, K Vanneste, F Murat, Soler M, San Clemente H , Saidi N, Cassan-Wang H, Dunand C, Hefer C, Bornberg-Bauer E, Kersting A, Vinnig K, Amarasinghe V, Ranik M, Naithani S, Elser J, Boyd A, Liston A, Spatafora J, Dharmawardhana P, Raja R, Sullivan C, Romanel E, Alves-Ferreira A, Külheim C, Foley W, Carocha V, Paiva J, Kudrna D, Brommonschenkel S, Pasquali G, Byrne M, Rigault P, Tibbits J, Spokevicius A, Jones R, Steane D, Vaillancourt R, Potts B, Barry K, Pappas G Jr, Strauss S, Jaiswal P, Grima-Pettenati J, Salse J, Van de Peer Y, Rokhsar D, Schmutz J, Van der Merwe K, Singh P, Joubert F (2014) The genome of Eucalyptus grandis – a global tree for fiber and energy Nature 510(7505), 356–362 doi:10.1038/nature13308
Wisniewski M, Nassuth A, Teulières C, Marque C, Rowland J, Cao PB, Brown A (2014) Genomics of cold hardiness in woody plants. Crit Rev Plant Sci 33 : 92-124.
Courtial A, Méchin V, Reymond M, Grima-Pettenati J, Barrière Y (2014). Colocalizations between several QTLs for cell wall degradability and composition in the F288 x F271 early maize RIL progeny raise the question of the nature of the possible underlying determinants and breeding targets for biofuel capacity. BioEnergy Res 7 : 142-156.
De la Torre F, Rodríguez R, Gago J, Villar B, Álvarez-Otero R, Grima-Pettenati J, Gallego PP (2014) Genetic transformation of Eucalyptus globulus : advantages of using the vascular-specific EgCCR over the constitutive CaMV35S promoter. Plant Cell Tiss Organ Cult 117 : 77-84 .
Camargo ELO, Nascimento LC, Soler M, Salazar MM, Lepikson-Neto J, Marques WL, Alves A, Teixeira PJPL, Mieczkowski P, Carazzolle MF, Martinez Y, Deckmann AC, Rodrigues JC, Grima-Pettenati J, Pereira G. (2014). Contrasting nitrogen fertilization treatments impact xylem gene expression and secondary cell wall lignification in Eucalyptus. BMC Plant Biol 14:256
2013
Keller G, Cao PB, San Clemente H, El Kayal W, Marque C, and Teulières C. (2013) Transcript profiling combined with functional annotation of 2,662 ESTs provides a molecular picture of Eucalyptus gunnii cold acclimation. trees, DOI : 10.1007/s00468-013-0918-5
Cassan-Wang H, Goue N, Saidi MN, Legay S, Sivadon P, Goffner D, and Grima-Pettenati J. 2013. Identification of novel transcription factors regulating secondary cell wall formation in Arabidopsis. Front Plant Sci 4, 189.
Courtial A, Soler M, Chateigner-Boutin A, Reymond M, Méchin V, Wang H, Grima-Pettenati J, Barriere Y. (2013) Breeding grasses for capacity to biofuel production or silage feeding value : An updated list of genes involved in maize secondary cell wall biosynthesis and assembly. Maydica 58 : 67-102.
Courtial A, Thomas J, Reymond M, Méchin V, Grima-Pettenati J, Barrière Y (2013) Targeted linkage map densification to improve cell wall related QTL detection and interpretation in maize. Theor Appl Genet 126 : 1151-1165.
2012
Cassan-Wang H, Soler M, Yu H, Camargo, ELO, Carocha V, Ladouce N, Savelli B, Paiva JaP, Leple JC, and Grima-Pettenati J 2012. Reference Genes for High-Throughput Quantitative Reverse Transcription-PCR Analysis of Gene Expression in Organs and Tissues of Eucalyptus Grown in Various Environmental Conditions. Plant and Cell Physiology 53 , 2101-16.
Amelot N, Dorlhac De Borne F, San Clemente H, Mazars C, Grima-Pettenati J, Briere C 2012. Transcriptome analysis of tobacco BY-2 cells elicited by cryptogein reveals new potential actors of calcium-dependent and calcium-independent plant defense pathways. Cell Calcium 51 , 117-30.
De Micco V, Ruel K, Joseleau JP, Grima-Pettenati J, Aronne G. (2012) Xylem anatomy and cell wall ultrastructure of Nicotianan tabacum L. after lignin genetic modification through transcriptional activator EgMYB2. IAWA Journal, Vol. 33 (3) 1–18
Azar S, Marque G, Clanet C, Marque C and Teulières C. (2012) Natural Variation in CBF Gene Sequence and Freezing Tolerance in Eucalyptus gunnii. ScienceMED, vol. 3, 259-265, Bologna
Courtial C, Jourda C, Arribat S, Balzergue S, Huguet S, Reymond M, Grima-Pettenati J, Barrière Y (2012) Comparative expression of cell wall related genes in four maize RILs and one parental line of variable lignin content and cell wall degradability. Maydica 57:56-74
Ouvrage international
Grima-Pettenati J, Soler M, Camargo E and Wang H. Transcriptional Regulation of the Lignin Biosynthetic Pathway Revisited : New Players and Insights. In Lise Jouanin and Catherine Lapierre, editors : Advances in Botanical Research, Vol. 61, Burlington : Academic Press, 2012, pp. 173-218. ISBN : 978-0-12-416023-1
2011
Amelot N, Carrouche A, Danoun S, Bourque S, Haiech J, Pugin A, Ranjeva R, Grima-Pettenati J, Mazars C, Briere C (2011) Cryptogein, a fungal elicitor, remodels the phenylpropanoid metabolism of tobacco cell suspension cultures in a calcium-dependent manner. Plant Cell Environ 34 : 149-161
Navarro M, Ayax C, Martinez Y, Laur J, El Kayal W, Marque C, Teulieres C (2011) Two EguCBF1 genes overexpressed in Eucalyptus display a different impact on stress tolerance and plant development. Plant Biotechnol J 9 : 50-63
Paiva JAP, Prat E, Vautrin S, Santos MD, San-Clemente H, Brommonschenkel S, Fonseca PGS, Grattapaglia D, Song XA, Ammiraju JSS, Kudrna D, Wing RA, Freitas AT, Berges H, Grima-Pettenati J (2011) Advancing Eucalyptus genomics : identification and sequencing of lignin biosynthesis genes from deep-coverage BAC libraries. BMC Genomics 12 : 137
Gago J, Grima-Pettenati J, Gallego PP (2011) Vascular-specific expression of GUS and GFP reporter genes in transgenic grapevine (Vitis vinifera L. cv. Albarino) conferred by the EgCCR promoter of Eucalyptus gunnii. Plant Physiology and Biochemistry 49 : 413-419
Gion JM, Carouche A, Deweer S, Bedon F, Pichavant F, Charpentier JP, Bailleres H, Rozenberg P, Carocha V, Ognouabi N, Verhaegen D, Grima-Pettenati J, Vigneron P, Plomion C Comprehensive genetic dissection of wood properties in a widely-grown tropical tree : Eucalyptus. Bmc Genomics 12 : 301
Wang H, Soler M, Yu H, Camargo E, San Clemente H, Savelli B, Ladouce N, Paiva J, Grima-Pettenati1 J (2011) Master regulators of wood formation in Eucalyptus. Wang et al. BMC Proceedings 2011, 5 (Suppl 7) : P110. http://www.biomedcentral.com/1753-6…
Camargo E, Costa L, Soler M, Salazar M, Lepikson J, Gonçalves D, Marques W, Carazzolle M, Martinez Y, Grima-Pettenati J, Pereira G (2011) Effects of nitrogen fertilization on global xylem transcript profiling of Eucalyptus urophylla x grandis evaluated by RNA-seq technology. BMC Proceedings, 5(Suppl 7):P106 http://www.biomedcentral.com/1753-6…
Paiva J, Rodrigues JC, Fevereiro P, Neves L, Araújo C, Marques C, Freitas AT, Bergès H, Grima-Pettenati J (2011) Building up resources and knowledge to unravel transcriptomics dynamics underlying Eucalyptus globulus xylogenesis. BMC Proceedings, 5(Suppl 7):O52
2010
Bedon F, Bomal C, Caron S, Levasseur C, Boyle B, Mansfield SD, Schmidt A, Gershenzon J, Grima-Pettenati J, Seguin A, MacKay J (2010) Subgroup 4 R2R3-MYBs in conifer trees : gene family expansion and contribution to the isoprenoid- and flavonoid-oriented responses. J Exp Bot 61 : 3847-3864
Legay S, Sivadon P, Blervacq AS, Pavy N, Baghdady A, Tremblay L, Levasseur C, Ladouce N, Lapierre C, Seguin A, Hawkins S, Mackay J, Grima-Pettenati J (2010) EgMYB1, an R2R3 MYB transcription factor from eucalyptus negatively regulates secondary cell wall formation in Arabidopsis and poplar. New Phytologist 188 : 774-786
Rahantamalala A, Rech P, Martinez Y, Chaubet-Gigot N, Grima-Pettenati J, Pacquit V (2010) Coordinated transcriptional regulation of two key genes in the lignin branch pathway—CAD and CCR—is mediated through MYB- binding sites. BMC Plant Biol 10 : 130
Saidi MN, Ladouce N, Hadhri R, Grima-Pettenati J, Drira N, Gargouri-Bouzid R (2010) Identification and characterization of differentially expressed ESTs in date palm leaves affected by brittle leaf disease. Plant Science 179 : 325-332
2009
Navarro M, Marque G, Ayax C, Keller G, Borges JP, Marque C, Teulieres C (2009) Complementary regulation of four Eucalyptus CBF genes under various cold conditions. J Exp Bot 60 : 2713-2724
Keller G, Marchal T, SanClemente H, Navarro M, Ladouce N, Wincker P, Couloux A, Teulières C and Marque C (2009) Development and functional annotation of a 11303 EST collection from Eucalyptus for studies of cold tolerance, Tree genetics and genome 5 : 317-327.
Bedon F, Levasseur C, Grima-Pettenati J, Seguin A, MacKay J (2009) Sequence analysis and functional characterization of the promoter of the Picea glauca Cinnamyl Alcohol Dehydrogenase gene in transgenic white spruce plants. Plant Cell Rep 28 : 787-800
Foucart C, Jauneau A, Gion JM, Amelot N, Martinez Y, Panegos P, Grima-Pettenati J, Sivadon P (2009) Overexpression of EgROP1, a Eucalyptus vascular-expressed Rac-like small GTPase, affects secondary xylem formation in Arabidopsis thaliana. New Phytol 183 : 1014-1029
Rengel D, San Clemente H, Servant F, Ladouce N, Paux E, Wincker P, Couloux A, Sivadon P, Grima-Pettenati J (2009) A new genomic resource dedicated to wood formation in Eucalyptus. BMC Plant Biol 9 : 36
2008
Bomal C, Bedon F, Caron S, Mansfield SD, Levasseur C, Cooke JE, Blais S, Tremblay L, Morency MJ, Pavy N, Grima-Pettenati J, Seguin A, Mackay J (2008) Involvement of Pinus taeda MYB1 and MYB8 in phenylpropanoid metabolism and secondary cell wall biogenesis : a comparative in planta analysis. J Exp Bot 59 : 3925-3939
Wadenback J, von Arnold S, Egertsdotter U, Walter MH, Grima-Pettenati J, Goffner D, Gellerstedt G, Gullion T, Clapham D (2008) Lignin biosynthesis in transgenic Norway spruce plants harboring an antisense construct for cinnamoyl CoA reductase (CCR). Transgenic Res 17 : 379-392
2007
Bedon F, Grima-Pettenati J, Mackay J (2007) Conifer R2R3-MYB transcription factors : sequence analyses and gene expression in wood-forming tissues of white spruce (Picea glauca). BMC Plant Biol 7 : 17
Legay S, Lacombe E, Goicoechea M, Briere C, Seguin A, Mackay J, Grima-Pettenati J (2007) Molecular characterization of EgMYB1, a putative transcriptional repressor of the lignin biosynthetic pathway. Plant Science 173 : 542-549
Leple JC, Dauwe R, Morreel K, Storme V, Lapierre C, Pollet B, Naumann A, Kang KY, Kim H, Ruel K, Lefebvre A, Joseleau JP, Grima-Pettenati J, De Rycke R, Andersson-Gunneras S, Erban A, Fehrle I, Petit-Conil M, Kopka J, Polle A, Messens E, Sundberg B, Mansfield SD, Ralph J, Pilate G, Boerjan W (2007) Downregulation of cinnamoyl-coenzyme A reductase in poplar : multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 19 : 3669-3691
2020
- Dai Y, Hu G, Dupas A, Medina L, Blandels N, Clemente HS, Ladouce N, Badawi M, Hernandez-Raquet G, Mounet F, Grima-Pettenati J, Cassan-Wang H (2020) Implementing the CRISPR/Cas9 Technology in Eucalyptus Hairy Roots Using Wood-Related Genes. Int J Mol Sci 21
- Cao PB, Ployet R, Nguyen C, Dupas A, Ladouce N, Martinez Y, Grima-Pettenati J, Marque C, Mounet F, Teulieres C (2020) Wood Architecture and Composition Are Deeply Remodeled in Frost Sensitive Eucalyptus Overexpressing CBF/DREB1 Transcription Factors. Int J Mol Sci 21
2019
- Gomes C, Dupas A, Pagano A, Grima-Pettenati J, Paiva JAP (2019) Hairy Root Transformation: A Useful Tool to Explore Gene Function and Expression in Salix spp. Recalcitrant to Transformation. Front Plant Sci 10: 1427
- Ployet R, Venezio-Labate MT, Regiani Catalidi T, Christina M, Morel M, San Clemente H, Denis M, Favreau B, Tomazello Filho M, Laclau JP, Labate CA, Chaix G, Grima-Pettenati J and F Mounet (2019). A system biology viw of wood formation in Eucalyptus grandis trees submitted to different potassium and water regimes. New Phytol .
- Camargo E, Ployet R, Cassan-Wang H, Mounet F, Grima-Pettenati J. 2019. Digging in wood: New insights in the regulation of wood formation in tree species. Advances in Botanical Research 89: 201-233.
- Hussey SG, Grima-Pettenati J, Myburg AA, Mizrachi E, Brady SM, Yoshikuni Y, Deutsch S. 2019. A Standardized Synthetic Eucalyptus Transcription Factor and Promoter Panel for Re-engineering Secondary Cell Wall Regulation in Biomass and Bioenergy Crops. Acs Synthetic Biology 8(2): 463-465.
- Lin Z, Long JM, Yin Q, Wang B, Li HL, Luo JZ, Wang HC, Wu AM. 2019. Identification of novel lncRNAs in Eucalyptus grandis. Industrial Crops and Products 129: 309-317.
- Tavares EQP, De Souza AP, Romim GH, Grandis A, Plasencia A, Gaiarsa JW, Grima-Pettenati J, de Setta N, Van Sluys MA, Buckeridge MS. 2019. The control of endopolygalacturonase expression by the sugarcane RAV transcription factor during aerenchyma formation. Journal of Experimental Botany 70(2): 497-506.
- Xu C, Shen Y, He F, Fu X, Yu H, Lu W, Li Y, Li C, Fan D, Wang HC, Luo K. 2018. Auxin-mediated Aux/IAA-ARF-HB signaling cascade regulates secondary xylem development in Populus. New Phytol.
2018
- Plomion C, Aury JM, Amselem J, Leroy T, Murat F, Duplessis S, Faye S, Francillonne N, Labadie K, Le Provost G, Lesur I, Bartholome J, Faivre-Rampant P, Kohler A, Leple JC, Chantret N, Chen J, Dievart A, Alaeitabar T, Barbe V, Belser C, Berges H, Bodenes C, Bogeat-Triboulot MB, Bouffaud ML, Brachi B, Chancerel E, Cohen D, Couloux A, Da Silva C, Dossat C, Ehrenmann F, Gaspin C, Grima-Pettenati J, Guichoux E, Hecker A, Herrmann S, Hugueney P, Hummel I, Klopp C, Lalanne C, Lascoux M, Lasserre E, Lemainque A, Desprez-Loustau ML, Luyten I, Madoui MA, Mangenot S, Marchal C, Maumus F, Mercier J, Michotey C, Panaud O, Picault N, Rouhier N, Rue O, Rustenholz C, Salin F, Soler M, Tarkka M, Velt A, Zanne AE, Martin F, Wincker P, Quesneville H, Kremer A, Salse J. 2018. Oak genome reveals facets of long lifespan. Nat Plants 4(7): 440-452
2017
- Ployet R, Soler M, Carocha V, Ladouce N, Alves A, Rodrigues JC, Harvengt L, Marque C, Teulières C, Grima-Pettenati J, Mounet F (2017) Long cold exposure induces transcriptional and biochemical remodelling of xylem secondary cell wall in Eucalyptus. Tree Physiol: 1-14
- Nguyen HC, Cao PB, San Clemente H, Ployet R, Mounet F, Ladouce N, Harvengt L, Marque C, Teulieres C (2017) Special trends in CBF and DREB2 groups in Eucalyptus gunnii vs Eucalyptus grandis suggest that CBF are master players in the trade-off between growth and stress resistance. Physiol Plant 159: 445-467
2016
- Barrière Y, Courtial A, Chateigner-Boutin AL, Denoue D, Grima-Pettenati J (2016) Breeding maize for silage and biofuel production, an illustration of a step forward with the genome sequence. Plant Science 242: 310-329
- Plasencia A, Soler M, Dupas A, Ladouce N, Silva-Martins G, Martinez Y, Lapierre C, Franche C, Truchet I, Grima-Pettenati J (2016) Eucalyptus hairy roots, a fast, efficient and versatile tool to explore function and expression of genes involved in wood formation. Plant Biotechnology Journal 14: 1381-1393
- Sixto H, Gonzalez-Gonzalez BD, Molina-Rueda JJ, Garrido-Aranda A, Sanchez MM, Lopez G, Gallardo F, Canellas I, Mounet F, Grima-Pettenati J, Canton F (2016) Eucalyptus spp. and Populus spp. coping with salinity stress: an approach on growth, physiological and molecular features in the context of short rotation coppice (SRC). Trees-Structure and Function 30: 1873-1891
- Soler M, Plasencia A, Larbat R, Pouzet C, Jauneau A, Rivas S, Pesquet E, Lapierre C, Truchet I, Grima-Pettenati J (2017) The Eucalyptus linker histone variant EgH1.3 cooperates with the transcription factor EgMYB1 to control lignin biosynthesis during wood formation. New Phytologist 213: 287-299
- Soler M, Plasencia A, Lepikson-Neto J, Camargo ELO, Dupas A, Ladouce N, Pesquet E, Mounet F, Larbat R, Grima-Pettenati J (2016) The Woody-Preferential Gene EgMYB88 Regulates the Biosynthesis of Phenylpropanoid-Derived Compounds in Wood. Frontiers in Plant Science 7
Theses
Thèses en cours
Ines HADJ-BACHIR (2019-2022) Crosstalk between cold signaling involving CBF transcription factors and regulation of wood formation in Eucalyptus
*******************
Thèses achevées
Ying Dai (2016-2020) Mise au point de la technologie CRISPR/Cas9 dans des racines « hairy root » d’Eucalyptus grandis et caractérisation fonctionnelle de facteurs de transcription de la signalisation auxinique potentiellement impliqués dans la formation du bois
Sahar Azar (2009-2012) Développement de marqueurs moléculaires SNP sur les gènes de facteurs de transcription CBF en vue de l’amélioration de la tolérance au froid de l’Eucalyptus
Camille Foucart (2006) « Recherche et caractérisation fonctionnelle de gènes régulateurs impliqués dans la xylogenèse chez Eucalyptus gunnii ».
Guylaine Keller (2006) « Analyse du transcriptome de l’Eucalyptus pendant l’acclimatation au froid : Recherche de gènes candidats de tolérance au gel »
Walid El Kayal (2004) « Réponses aux stress abiotiques chez Eucalyptus gunnii : analyse globale sur filtres hautes densité et caractérisation moléculaire du gène Sxd1 (synthèse de la VitE) » .
Etienne Paux (2004) « Identification de gènes-candidats impliques dans la formation du xylème chez l’Eucalyptus ».
lrsv.gestion@univ-tlse3.fr.
Phone
Standard : +33 5.34.32.38.01
Fax : +33 5.34.32.38.02
Find us
24, chemin de Borde-Rouge.BP 42617 Auzeville.
31326, Castanet-Tolosan. FRANCE




