Our Team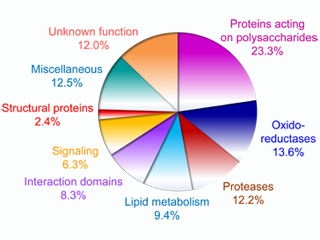
Vincent BURLAT
Professor - University Paul Sabatier-Toulouse 3
burlat@lrsv.ups-tlse.fr
Hervé CANUT
Researcher at CNRS
canut@lrsv.ups-tlse.fr

Josiane CHOURRE
Technician - University Paul Sabatier-Toulouse 3
chourre@lrsv.ups-tlse.fr
Laurent HOFFMANN
Assistant Professor - University Paul Sabatier-Toulouse 3
hoffmann@lrsv.ups-tlse.fr
David ROUJOL
Engineer - University Paul Sabatier-Toulouse 3
roujol@lrsv.ups-tlse.fr
Bastien DAUPHIN
PhD student - Université Paul Sabatier-Toulouse 3
bastien.dauphin@lrsv.ups-tlse.fr

Hasan KOLKAS
PhD student - Paul Sabatier - Toulouse 3 University
Sébastien VIUDES
PhD student - Paul Sabatier-Toulouse 3 University
sebastien.viudes@lrsv.ups-tlse.fr
Former members
Cécile ALBENNE (Assistant Professor UPS, 2005-2016)
Kevin BELLANDE (PhD student, 2015-2018)
Edith FRANCOZ (PhD student/ATER, 2012-2016)
May HIJAZI (PhD student, 2008-2011)
Muhammad IRSHAD (PhD student, 2005-2008)
Adélaïde JACQ (PhD student, 2013-2016)
Huan NGUYEN-KIM (PhD student, 2011-2015)
Jonathas PEREIRA DAS GRACAS (PhD student, Sao Paulo University, Brasil, 2017)
Rafael PONT-LEZICA (emeritus professor, UPS)
Research Topics
The plant cell wall is a dynamic structure playing important roles during growth, differentiation, during the interactions with pathogens and in response to environmental abiotic factors. The cell wall is both a source and a site of perception of signals. Our work aims at understanding the function of cell wall proteins during development. It requires (i) the identification of cell wall proteins and of the corresponding genes, (ii) the following of the dynamics of the cell wall proteome in various physiological conditions, and (iii) functional analyses.
Four research topics are presently worked out:
Plant cell wall proteomics
Contact: Elisabeth JAMET
For a long time considered as rigid envelopes around plant cells, cell walls now appear as dynamic compartments involved in many physiological processes such as developpement, signaling or defense against biotic and abiotic stresses. Plant cell walls are mainly composed of polysaccharides (about 90% in mass) but they also contain proteins. Their biochemical composition and the organization of their components change during development and in response to environmental factors. Both polysaccharides and proteins can be modified to allow the adaptation of cells. The present knowledge of the roles of cell wall proteins is still limited. The coming challenge is to decipher their function and to understand their interactions in muro.
The cell wall proteome of A. thaliana
Since the beginning of the 2000’s, the team has gained renown in plant cell wall proteomics and has got a leader position in this field. It has greatly contributed to enlarge the knowledge of the cell wall proteome of the model plant Arabidopsis thaliana by analyzing different plant materials using various strategies: cell suspension cultures (Borderies et al. 2003), rosette leaves (Boudart et al. 2005, Hervé et al. 2016), etiolated hypocotyls (Feiz et al. 2006, Irshad et al. 2008), roots (Nguyen-Kim et al. 2016) and secretome of etiolated seedlings in liquid culture medium (Charmont et al. 2005). The glycoproteome of etiolated hypocotyls has been studied in detail, showing the importance of N-glycosylated cell wall proteins (Zhang, Giboulot et al. 2011). All these data, combined to others has brought to 763 the number of identified cell wall proteins of A. thaliana, that is more than one third of the expected total number of cell wall proteins (Albenne et al. 2013, Nguyen-Kim et al. 2016, Hervé et al. 2016).
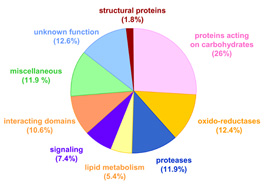
Systematic cell wall proteomics has been extended to other plant species such as Brachypodium distachyon, Saccharum officinarum and Triticum aestivum. The cell wall proteome of B. distachyon is the best described among monocots with 596 proteins identified (Douché et al. 2013, Francin-Allami et al. 2015, Francin-Allami et al. 2016). Cell wall proteomics data obtained by the team and other research groups have been collected in the WallProtDB database. This database presently comprises 3401 proteins from 12 plant species (Albenne et al. 2014, San Clemente and Jamet 2015). All the proteins have been re-annotated according to protein domain databases, thus facilitating the comparison between proteomes. For a clearer overview of cell wall proteomes, they have been grouped into nine functional classes (Jamet et al. 2008). WallProtDB has been referenced as a OMIC tool and at Database Commons.
The team has created an additional complementary bioinformatic tools to facilitate proteomic analyses. ProtAnnDB allows collecting predictions of sub-cellular localization and of functional domains obtained with online available software (San Clemente et al. 2009). It permits quick and efficient annotation of proteins identified by mass spectrometry and present cntains annotations of proteins from 18 plant species.
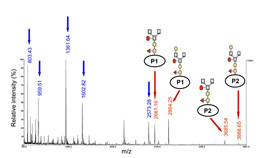
- Identification of N-glycopeptides of the At3g32980 class III peroxidase by MALDI-TOF MS
Our work also allowed to study post-translational modifications of structural proteins by the use of different approaches for protein separation and the use of mass spectrometry (Canut et al. 2016). We could localize modified amino acids not yet described, N– and O-glycosylations and protein maturation sites (Albenne et al. 2009; Zhang, Giboulot et al. 2011; Hijazi et al. 2012).
A systematic search for the presence of hydroxyproline residues (Hyp) in mass spectrometry data a allowed the identification of new protein families carrying this post-translational modification. A new proline hydroxylation code has been proposed (Canut et al. 2016, Duruflé et al. 2017). It allows predicting the position of Hyp residues in Pro-rich proteins (HRGPs), but remains inefficient in the case of other protein families.
Altogether, these proteomic studies have enabled the identification of cell wall proteins which functional analyses are underway through genetic approaches. We are presently particularly interested in Pro/Hyp-rich proteins, lectins, proteins involved in lipid metabolism and proteins of unknown function.
Structure-function relationships of cell wall proteins having domains interacting with cell wall components
Contact : Elisabeth JAMET
Besides sytematic proteomic approaches, the team is interested in elucidating the structure and the function of some cell wall proteins. During the last years, Arabidopsis thaliana DUF642 proteins and AGP31 (ArabinoGalactan Protein 31) encoded by At1g28290, have been particularly studied. These proteins have been identified as abundant proteins in cell walls of etiolated hypocotyls which undergo an important elongation within a few days.
The DUF642 (Domain of Unknown function 642, PF04862) protein family comprises 10 members in A. thaliana and is well represented in all the plant cell wall proteomes studied so far (see WallProtDB). These proteins comprise a N-terminal signal peptide and two sub-domains sharing conserved amino acids predicted to be folded as beta-sheets. A few members of the family contain a GPI anchor. Such proteins have been found in all the spermatophyte genomes analyzed (Vasquez-Lobo, Roujol et al. 2012). We could show that one DUF642 protein (At3g08030) interact with cellulose in vitro (Vasquez-Lobo, Roujol et al. 2012).
AGP31 comprise a N-terminal peptide signal, a short AGP domain, a His-rich domain, a central Pro-rich central domain as well as a C-terminal domain called PAC (PRP-AGP-copntaining Cys) domain. This multi-domain organization is unique in A. thaliana and has retained our attention.
The combination of several mass spectrometry techniques associated to biochemical characterization of the protein has allowed to show the presence of Hyp residues and of Ara/Gal O-glycans in the Pro-rich domain (Hijazi et al. 2012). Several isoforms of AGP31 have been found, differing in the type and size of O-glycans or in the size of the polypeptide moiety. In particular, the AGP domain carries arabinogalactan (AG) chains which structures remain to be elucidated. Besides, in vitro interactions have been demonstrated between the PAC domain of AGP31 and galactans also found as lateral chains of type I rhamnogalacturonans (RG I) (Hijazi et al. 2014). In addition, AGP31 can interact with itself probably through the O-glycans of its Pro-rich domain. Finally, the O-glycans of the Pro-rich domain are recognized by a peanut agglutinin (PNA), suggesting possible interactions with cel wall lectins.
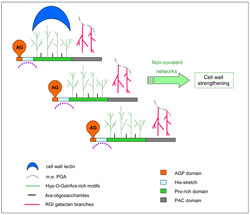
Altogether, these data allowed to propose a model of non covalent interactions putting AGP31 in the middle of a complex supremolecular scaffold, thus suggesting a role of cell wall organizer for AGP31. These results bring new ideas about the architecture of plant cell walls and enlighten the central role of O-glycoproteins in cell wall structure (Hijazi et al. 2014).
Lectin receptor kinases and Cell wall-sensing mechanisms
Contact : Hervé CANUT
Our group investigates communication between the cell wall and the cytoplasm of plants. One focus is to decipher the functions of lectins in the model plant Arabidopsis thaliana.
Lectins are widespread proteins in all kingdoms of life. They are able to bind mono-or oligosaccharides in a reversible way and to decipher the information contained in these complex structures. Lectins, mainly originating from plants, find large applications in bioscience and biomedicine. From the original applications in blood banks, recent progress has been made in unravelling their effects, often beneficial, on tumours and in cancer therapy. Also, lectins are fundamental to plant life and play important roles in cell communication, development and natural defence strategies.
Cell walls are complex structures made of cellulose, hemicelluloses, pectins and proteins secreted by the cell, so setting up a rigid and continuous structure within the plant tissues. Cell walls are dynamic structures, sources of chemical or mechanical signals. In A. thaliana, the lectins are present in the cell walls both as soluble proteins weakly bound to the wall (Lecs), and as chimeric proteins located at the plasma membrane: lectins are then the extracellular domain of lectin receptor kinases (LecRKs).
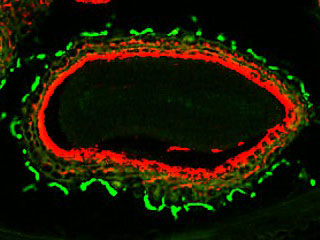
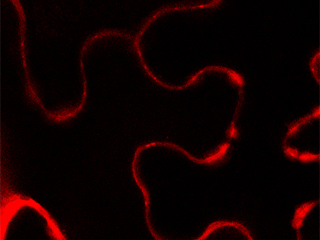
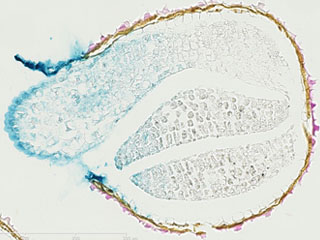
We have shown that, in A. thaliana, cell wall-plasma membrane contacts [1-white arrowheads] are set up by protein-protein interactions involving the recognition of the Arg-Gly-Asp (RGD) motif. LecRK-I.9 (At5g60300) is a candidate to be a linker between plasma membrane and cell wall and it belongs to the legume-type lectin family [2]. Recent results showed that LecRK-I.9 affects resistance of A. thaliana to Pseudomonas syringae pv. tomato (Pst), a bacterial pathogen and to Phytophthora infestans (Bouwmeester et al. 2011, Balagué et al. 2016). Besides, our analysis of the cell wall proteome of elongating cells has revealed several Lecs (Irshad et al. 2008). From amino acid sequences analysis, the legume lectin domains of both Lecs and LecRKs are closely related and form a cluster in phylogenic trees. Finally, LecRK-I.9 promoter-GUS reporter gene fusions have revealed that the LecRK-I.9 promoter activity is the highest in the apex of the main root and absent from the primordia of lateral roots [3].
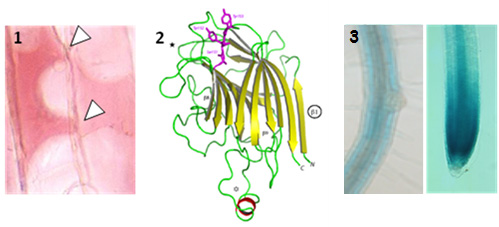
The aim of the project is to elucidate the role of Lecs/LecRKs in A. thaliana. A combination of biochemistry and functional genomics approaches will be carried out to identify the Lecs/LecRKs extracellular ligands and to unravel the functions of lectins. We assume that Lecs/LecRKs are involved in the detection of pathogens, but also in the assembly and the remodelling of the cell wall polysaccharides. In particular, Lecs/LecRKs could act as sensors and be sentinels reporting the cell wall integrity.
Functional analysis of CIII peroxidases during Arabidopsis thaliana development
Contact : Christophe DUNAND et Philippe RANOCHA (Evolution and Expression of Peroxidases Team),Vincent Burlat (Cell wall proteins and Development team)
The 73 class III peroxidase (Prx) genes d’Arabidopsis thaliana show specific expression profiles and the corresponding proteins are predicted to be localized in the cell wall or in the vacuole, depending on the presence/absence of a C-terminal vacuole sorting amino acid motif (Francoz et al. 2014). Recent studies have suggested that a given Prx could co-localize with its substrate, thus allowing a control of its activity in so-called cell wall micro-domains. As a consequence, the cell wall could be relaxed or reinforced.
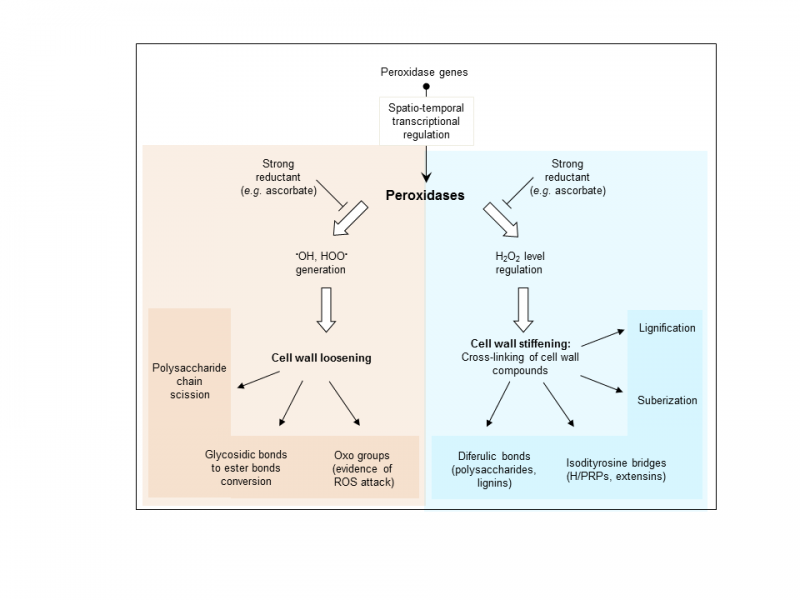
In the frame of a transverse project involving both the Evolution and Expression of Peroxidases and the Cell wall proteins and Development teams, we are presently studying the role of cell wall Prxs during two developmental processes leading to the formation of the seed and its germination. The selection of candidates of interest is based on the fine mining of transcriptomics data and on results of in situ RNA hybridizations (Francoz et al. 2016). The functional analysis of these genes is performed by reverse genetics coupled to micro-phenotyping.
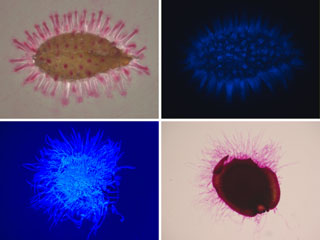
Control of the dynamics of cell walls of seed mucilage secretory cells (MSCs)
During embryogenesis, the five cell layers forming the seed teguments differentiate, the outermost epidermal layer showing a remarkable cell wall dynamics. Indeed, in addition to the primary cell wall, these cells synthesize an internal cell wall called the volcano-shaped columella and produce a polysaccharidic mucilage compressed between the tangential primary cell wall and this columella. This mucilage plays a role only at the time of seed imbibition several weeks to several years after the end of seed dessication. The reinforcement or relaxation zones which are localized in the different kinds of cell walls lead to a polarized rupture of the external cell wall under the pressure of the rehydrated mucilage. The germination process is then facilitated. Presently, we are looking for the function of several Prxs during this developmental process.
Control of the rupture of the mycropylar endosperm during germination
Germination is controled by external factors such as temperature, water availability, light and hormones. Two main steps of germination are the rupture of seed teguments, followed by that of the micropylar endosperm favoring the radicle emergence. Recently, the role of active oxygen species (ROS) as secondary messengers has been demonstrated during this process (Lariguet et al. 2013). It has been shown that ROS are released before the micropylar endosperm rupture, that the production of ROS is required for germination, and that Prxs could be involved in the spatio-temporal regulation of ROS production. Presently, we are studying several Prxs which could be involved in the control of micropylar endosperm rupture.
The Cell wall proteins and Development team has organized:
- The meeting of the French Cell Wall Network, in 2008 .

- The annual meeting of the French Green Proteomics Network (Protéome vert), in 2011

- The mixOmics Summer School in the frame of COST FA1306 The quest for tolerant varieties: phenotyping at plant and cellular level, in 2016.
Elisabeth JAMET has been an invited guest co-editor together with Els VAN DAMME, Nausicaa LANNOO and Cécile ALBENNE of an e-book entitled Plant Glycobiology – A sweet world of lectins, glycoproteins, glycolipids and glycans in the frame of the Research Topics of Frontiers in Plant Science (2015). She has also be an invited co-editor of the Plant proteomics 2017 Special Issue of Proteomes together with Véronique SANTONI. She has also been guest co-editor (i) with Christophe DUNAND of the Special Issue of International Journal of Molecular Sciences (IJMS) which will be devoted to Plant cell wall proteins and Development and (ii) with with Chistophe DUNAND and Zoë POPPERof a Special Topic of Frontiers in Plant Science dealing with Co-evolution of plant cell wall polymers together.
E JAMET is presently co-guest editor of a Special issue of Cells entitled Research on Plant cell wall biology which is presently open for manuscript submission.
Publications
Althiab-Almasud R, Chen Y, Maza E, Djari A, Frasse P, Mollet JC, Mazars C, Jamet E, Chervin C (2021) Ethylene signaling modulates tomato pollen tube growth, through modifications of cell wall remodeling and calcium gradient. Plant J ( (read)
Douché T, Valot B, Balliau T, Zivy M, San Clemente H, Jamet E (2021). Cell wall proteomic datasets of stems and leaves of Brachypodium distachyon. Data Brief 35: 106818 (read)
2020
Bellande K, Lalo A, Ligat L, Roujol D, Jamet E, Canut H (2020) Recombinant N-glycosylation isoforms of Legume lectins: production and purification from Nicotiana benthamiana leaves following RuBisCO depletion. Plant Physiol Biochem 157: 441-452 (read)
Cherkaoui M, Lollier V, Geairon A, Bouder A, Larré C, Rogniaux H, Jamet E, Guillon F, Francin-Allami M (2020) Cell wall proteome of wheat grain endosperm and outer layers at two key stages of early development. Int J Mol Sci 21: 239 (read)
Duruflé H, Ranocha P, Balliau T, Zivy M, Albenne C, Burlat V, Déjean S, Jamet E, Dunand C (2020) An integrative study showing the adaptation to sub-optimal growth conditions of natural populations of Arabidopsis thaliana: a focus on cell wall changes. Cells 9: 2249 (read)
Duruflé H, Selmani M, Ranocha P, Jamet E, Dunand C, Déjean S (2020) A powerful framework for an integrative study with heterogeneous omics data: from univariate statistics to multi-block analysis. Brief Bioinformatics bbaa166 (read)
Roujol D, Hoffmann L, San Clemente H, Ritzenthaler C, Burlat V, Jamet E (2020) Plant cell wall proteomes : bioinformatics and cell biology tools to assess the bona fide cell wall localization of proteins. Methods Mol Biol 2149: 443-462 (read)
Viudes S, Burlat V, Dunand C (2020) Seed mucilage evolution: diverse molecular mechanisms generate versatile ecological functions for particular environments. Plant Cell Environ (doi.org/10.1111/pce.13827) (read)
2019
Berthold F, Roujol D, Hemmer C, Jamet E, Ritzenthaler C, Hoffmann L, Schmitt-Keichinger C (2019) Inside or outside? A new collection of Gateway vectors allowing plant protein subcellular localization or over-expression. Plasmid 105: 102436 (read)
Francoz E, Ranocha P, Dunand C, Burlat V (2019). Medium-throughput RNA in situ hybridization of serial sections from paraffin-embedded tissue microarrays. Methods Mol Biol, 1933: 99-130 (read)
Francoz E, Ranocha P, Le Ru A, Martinez Y, Fourquaux I, Jauneau A, Dunand C, Burlat (2019) Pectin demethylesterification generates platforms that anchor peroxidases to remodel plant cell wall domains. Dev Cell 48: 261-276 (read)
Duruflé H, Albenne C, Jamet E, Dunand C (2019) Phenotyping and cell wall polysaccharide composition of five Arabidopsis ecotypes grown at optimal or sub-optimal temperatures. Data brief 25: 104318 (read)
Duruflé H, Ranocha P, Balliau T, Dunand C, Jamet E (2019) Transcriptomic and cell wall proteomic datasets of rosettes and floral stems from five Arabidopsis thaliana ecotypes grown at sub-optimal temperature. Data Brief 27: 104581 (read)
Duruflé H, Selmani M, Ranocha P, Jamet E, Dunand C, Déjean S (2019) A powerful framework for an integrative study with heterogeneous omics data: from univariate statistics to multi-block analysis. bioRxiv (
2018
Calderan-Rodrigues MJ, Fonseca JG, San Clemente H, Labate CA, Jamet E (2018) Glycoside hydrolases in plant cell wall proteomes: predicting functions that could be relevant for improving biomass transformation processes. Advances in Biofuels and Bioenergy (M Nageshwara-Rao and J Soniji eds), InTechOpen access Publisher, Croatia, pp 165-182 (read)
Cherkaoui M, Geairon A, Lollier V, San Clemente H, Larré C, Rogniaux H, Jamet E, Guillon F, Francin-Allami M. (2018) Cell wall proteome investigation of bread wheat (Triticum aestivum) developing grain in endosperm and outer layers. Proteomics 18: 180086 (read)
Fonseca JG, Calderan-Rodrigues MJ, de Moraes FE, Cataldi TR, Jamet E, Labate CA (2018) Cell wall proteome of sugarcane young and mature leaves and stems. Proteomics 18: 1700129 (read)
Jacq A, Burlat V, Jamet E (2018) Plant cell wall proteomics as a strategy to reveal candidate proteins involved in extracellular lipid metabolism. Curr Protein Pept Sci 19: 190-199 (read)
Jamet E, Santoni V (2018) Editorial for Special Issue: 2017 Plant Proteomics. Proteomes 6: 28 (read)
2017
Badruna L, Burlat V, Montanier C (2017) CBMs as probes to explore plant cell wall heterogeneity using immunocytochemistry. Methods Mol Biol 1588 : 181-197 (read)
Balagué C, Gouget A, Bouchez O, Souriac C, Haget N, Boutet-Mercey S, Govers F, Roby D, Canut H (2016) The Arabidopsis thaliana lectin receptor kinase LecRK-I.9 is required for full resistance to Pseudomonas syringae and affects jasmonate signalling. Mol Plant Pathol 18 : 937-948 (read)
Bellande K, Bono JJ, Savelli B, Jamet E, Canut H (2017) Plant lectins and lectin receptor-like kinases : How do they sense the outside ? Int J Mol Sci 18 : 1164 (read)
Calderan-Rodrigues MJ, Fonseca JG, Labate CA, Jamet E (2017) Cell wall proteomics as a means to identify target genes to improve second generation biofuel production. In Frontiers in Bioenergy and Biofuels (E Jacob-Lopes and L Queiroz Zepka ed), InTech Open access Publisher, Croatia, pp 5-24 (read)
Canut H, Albenne C, Jamet E (2017) Isolation of the cell wall. Methods Mol Biol 1511 : 171-185 (read)
Cosio C, Ranocha P, Francoz E, Burlat V, Zheng Y, Perry SE, Ripoll JJ, Yanofsky M, Dunand C (2017) The class III peroxidase PRX17 is a direct target of the MADS-box transcription factor AGAMOUS-LIKE15 (AGL15) and participates in lignified tissue formation. New Phytol 213 : 250-263 (read)
Duruflé H, Hervé V, Balliau T, Zivy M, Dunand C, Jamet E (2017) Proline hydroxylation in cell wall proteins : is it yet possible to define rules ? Front Plant Sci 8: 1802 (read)
Duruflé H, Hervé V, Ranocha P, Balliau T, Zivy M, Chourré J, San Clemente, H, Burlat V, Albenne C, Déjean S, Jamet E, Dunand C (2017) Cell wall modifications of two Arabidopsis thaliana ecotypes, Col and Sha, in response to sub-optimal growth conditions : an integrative study. Plant Sci 263 : 183-193 (read)
Duruflé H, San Clemente H, Balliau T, Zivy M, Dunand C, Jamet E (2017) Cell wall proteome analysis of Arabidopsis thaliana mature stems. Proteomics 17, 8, 1600449 (read)
Jacq A, Pernot C, Martinez Y, Domergue F, Payré B, Jamet E, Burlat V, Pacquit V (2017) The Arabidopsis Lipid Transfer Protein 2 (AtLTP2) is involved in cuticle-cell wall interface integrity and in etiolated hypocotyl permeability. Front Plant Sci 8 : 263 (read)
Jamet E (2017) Plant cell wall proteomics : An assessment twenty years after launching. Bioinform. Proteomics Open Access J 1: 000107 (read)
Regente M, Pinedo M, San Clemente H, Balliau T, Jamet E, de la Canal L (2017) Plant extracellular vesicles are incorporated by cells of a fungal pathogen and inhibit its growth. J Exp Bot 20: 5485-5496 (read)
Sibout R, Proost S, Hansen B, Vaid N, Giorgi F, Ho-Yue-Kuang S, Legée F, Cezart L, Bouchabké-Coussa O, Soulhat C, Provart N, Pasha A, Le Bris P, Roujol D, Höfte H, Jamet E, Lapierre C, Persson S, Mutwil M (2017) Expression atlas and comparative co-expression network analyses reveal important genes involved in the formation of lignified cell wall in Brachypodium distachyon. New Phytol 215: 1009-1025 (read)
2016
Calderan-Rodrigues MJ, Jamet E, Douché T, Rodrigues Bonassi MB, Regiani Cataldi T, Fonseca Guimarães J, San Clemente H, Pont-Lezica R, Labate CA (2016) Cell wall proteome of sugarcane stems : comparison of a destructive and a non-destructive extraction method showed differences in glycoside hydrolases and peroxidases. BMC Plant Biol 16 : 14 (read)
Canut H, Albenne C, Jamet E (2016) Post-translational modifications of plant cell wall proteins and peptides : A survey from a proteomics point of view. Biochim Biophys Acta 1864 : 983-990 (read)
Francin-Allami M, Lollier V, Pavlovic M, San Clemente H, Rogniaux H, Jamet E, Guillon F, Larré C (2016) Understanding the remodelling of cell walls during Brachypodium distachyon grain development through a sub-cellular quantitative proteomic approach. Proteomes 4 : 21 (read)
Francoz E, Ranocha P, Pernot C, Le Ru, A, Pacquit V, Dunand C, Burlat V (2016) Complementarity of medium-throughput in situ RNA hybridization and tissue-specific transcriptomics : case study of Arabidopsis seed development kinetics. Sci Rep 6:24644 (read)
Hervé V, Duruflé H, San Clemente H, Albenne C, Balliau T, Zivy M, Dunand C, Jamet E (2016) An enlarged cell wall proteome of Arabidopsis thaliana rosettes. Proteomics 16 : 3183-3187 (read)
Nguyen-Kim H, San Clemente H, Balliau T, Zivy M, Dunand C, Albenne C, Jamet E (2016) Arabidopsis thaliana root cell wall proteomics : increasing the proteome coverage using a combinatorial peptide ligand library and description of unexpected Hyp in peroxidase amino acid sequences. Proteomics 16 : 491-503 (read)
2015
Dupoiron S, Zischek C, Ligat L, Carbonne J, Boulanger A, Dugé de Bernonville T, Lautier M, Rival P, Arlat M, Jamet E, Lauber E, Albenne C (2015) The N-glycan cluster from Xanthomonas campestris pv. campestris : a toolbox for sequential plant N-glycan processing. J Biol Chem 290 : 6022-6036 (read)
Francin-Allami M, Merah K, Albenne C, Rogniaux H, Pavlovic M, Lollier V, Sibout R, Guillon F, Jamet E, Larré C (2015) Cell wall proteomics of Brachypodium distachyon grains : A focus on cell wall remodeling proteins. Proteomics 15 : 2296-2306 (read)
Francoz E, Ranocha P, Burlat V, Dunand C (2015) Arabidopsis seed mucilage 1 secretory cells : Regulation and dynamics. Trends Plant Sci 20 : 515-524 (read)
Francoz E, Ranocha P, Nguyen-Kim H, Jamet E, Burlat V, Dunand C (2015) Roles of cell wall peroxidases in plant development. Phytochemistry 112 : 15-21 (read)
Pont-Lezica RF (2015) Localizing proteins by tissue printing. Methods Mol Biol 1312 : 93-104 (read)
San Clemente H, Jamet E (2015) WallProtDB, a database resource for plant cell wall proteomics. Plant Methods 11 : 2 (read)
Sénéchal F, L’Enfant M, Domon JM, Rosiau E, Crépeau MJ, Surcouf O, Esquivel-Rodriguez J, Marcelo P, Mareck A, Guérineau F, Hyung-Rae K, Mravec J, Bonnin E, Jamet E, Kihara D, Lerouge P, Ralet MC, Pelloux J, Rayon C (2015) Tuning of pectin methylesterification : PECTIN METHYLESTERASE INHIBITOR 7 modulates the processive activity of co-expressed PECTIN MEHYLESTERASE 3 in a pH-dependent manner. J Biol Chem 290 : 23320-23335 (read)
2014
Albenne C, Canut H, Hoffmann L, Jamet E (2014) Plant cell wall proteins : a large body of data, but what about runaways ? Proteomes 2 : 224-242 (read)
Calderan-Rodrigues MJ, Jamet E, Calderan Rodrigues Bonassi MB, Guidetti-Gonzalez S, Carmanhanis Begossi A, Vaz Setem L, Franceschini LM, Guimarães Fonseca J, Labate CA (2014) Cell wall proteomics of sugarcane cell suspension cultures. Proteomics 14 : 738-749 (read)
Dugé de Bernonville T, Albenne C, Arlat M, Hoffmann L, Lauber E, Jamet E (2014) Xylem sap proteomics. Methods Mol Biol 1072 : 391-405 (read)
Hijazi M, Roujol D, Nguyen-Kim H, del Rocio Cisneros Castillo L, Saland E, Jamet E, Albenne C (2014) Arabinogalactan protein 31 (AGP31), a putative network-forming protein in Arabidopsis thaliana cell walls ? Ann Bot 114 : 1087-1097 (read)
Hijazi M, Velasquez MS, Jamet E, Estevez JM, Albenne C (2014) An update on post-translational modifications of hydroxyproline-rich glycoproteins : Towards a model highlighting their contribution to plant cell wall architecture. Front Plant Sci 5 : 395 (read)
Lannoo N, Van Damme EJM, Albenne C, Jamet E (2014) Plant Glycobiology – a diverse world of lectins, glycoproteins, glycolipids and glycans. Front Plant Sci 5 : 604 (read)
Ranocha P, Francoz E, Burlat V, Dunand C (2014) Expression of PRX36, PMEI6 and SBT1.7 is controlled by complex transcription factor regulatory networks for proper seed coat mucilage extrusion. Plant Signal Behav 9 : e977734 (read)
2013
Albenne C, Canut H, Jamet E (2013) Plant cell wall proteomics : the leadership of Arabidopsis thaliana. Front Plant Sci 4 : 111 (read)
Douché T, San Clemente H, Burlat V, Roujol D, Valot B, Zivy M, Pont-Lezica R, Jamet E (2013) Brachypodium distachyon as a model plant towards improved biofuel crops : Search for secreted proteins involved in biogenesis and disassembly of cell wall polymers. Proteomics 13 : 2438-2454 (read)
Mallèvre F, Roget A, Kervella Y, Ropartz D, Ralet M-C, Canut H, Minon T, Livache T (2013) Microwave heating for the rapid generation of glycosylhydrazides. Bioconjugate Chem 24 : 1264-1269 (read)
Simkin AJ, Miettinen K, Claudel P, Burlat V, Guirimand G, Courdavault V, Papon N, Meyer S, Godet S, St-Pierre B, Giglioli-Guivarc’h N, Fischer MJ, Memelink J, Clastre M (2013) Characterization of the plastidial geraniol synthase from Madagascar periwinkle which initiates the monoterpenoid branch of the alkaloid pathway in internal phloem associated parenchyma. Phytochemistry 85:36-43
2012
Geu-Flores F, Sherden NH, Courdavault V, Burlat V, Glenn WS, Wu C, Nims E, Cui Y, O’Connor SE (2012) An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature 492:138-142
Guirimand G, Guihur A, Phillips M, Oudin A, Glévarec G, Mahroug S, Melin C, Papon N, Clastre M, Giglioli-Guivarc’h N, St-Pierre B, Rodríguez-Concepción M, Burlat V, Courdavault V (2012) Triple subcellular targeting of isopentenyl diphosphate isomerases encoded by a single gene. Plant Signal Behav 7 : 1495-1497
Guirimand G, Guihur A, Phillips MA, Oudin A, Glévarec G, Melin C, Papon N, Clastre M, St-Pierre B, Rodríguez-Concepción M, Burlat V, Courdavault V (2012) A single gene encodes isopentenyl diphosphate isomerase isoforms targeted to plastids, mitochondria and peroxisomes in Catharanthus roseus. Plant Mol Biol 79 : 443-459
Guirimand G, Simkin AJ, Papon N, Besseau S, Burlat V, St-Pierre B, Giglioli-Guivarc’h N, Clastre M, Courdavault V (2012) Cycloheximide as a tool to investigate protein import in peroxisomes : a case study of the subcellular localization of isoprenoid biosynthetic enzymes. J Plant Physiol 169 : 825-829
Hijazi M, Durand J, Pichereaux C, Pont F, Jamet E, Albenne C (2012) Characterization of the ArabinoGalactan Protein 31 (AGP31) of Arabidopsis thaliana : new advances on the Hyp-O-glycosylation of the Pro-rich domain. J Biol Chem 287 : 9623-9632 (read)
Vázquez-Lobo A, Roujol D, Zuñiga-Sánchez E, Albenne C, Piñero D, Gamboa de Buen A, Jamet E (2012) The highly conserved spermatophyte cell wall DUF642 protein family : phylogeny and first evidence of interaction with cell wall polysaccharides in vitro. Mol Phylogenet Evol 63 : 510-520 (read)
2011
Bouwmeester K, de Sain M, Weide R, Gouget A, Klamer S, Canut H, Govers F (2011) The lectin receptor kinase LecRK-I.9 is a novel Phytophthora resistance component and a host target for a RXLR effector. PLoS Pathog 7 : e1001327 (read)
Guirimand G, Guihur A, Ginis O, Poutrain P, Héricourt F, Oudin A, Lanoue A, St-Pierre B, Burlat V, Courdavault V (2011) The subcellular organization of strictosidine biosynthesis in Catharanthus roseus epidermis highlights several trans-tonoplast translocations of intermediate metabolites. FEBS J 278 : 749-63
Guirimand G, Guihur A, Poutrain P, Héricourt F, Mahroug S, St-Pierre B, Burlat V, Courdavault V (2011) Spatial organization of the vindoline biosynthetic pathway in Catharanthus roseus. J Plant Physiol 6 : 549-557
Guénin S, Mareck A, Rayon C, Lamour R, Assoumou, Ndong Y, Domon JM, Sénéchal F, Fourneta F, Jamet E , Canut H, Percoco G, Mouille G, Rolland A, Rustérucci C, Guérineau F, Van Wuytswinkel O, Gillet F, Driouich A, Lerouge P, Gutierrez L, Pelloux J (2011) Identification of PME3 as a basic pectin methylesterase isoform involved in adventitious rooting in Arabidopsis. New Phytol 192 : 114-126 (read)
Jamet E (2011). Obituary : Rafael Pont-Lezica. Front Plant Sci 2:86
Ligat L, Lauber E, Albenne C, San Clemente H, Valot B, Zivy M, Pont-Lezica R, Arlat M, Jamet E (2011) Analysis of the xylem sap proteome of Brassica oleracea reveals a high content in secreted proteins. Proteomics 11 : 1798-1813 (read)
Poutrain P, Guirimand G, Mahroug S, Burlat V, Melin C, Ginis O, Oudin A, Giglioli-Guivarc’h N, Pichon O, Courdavault V (2011) Molecular cloning and characterisation of two calmodulin isoforms of the Madagascar periwinkle Catharanthus roseus. Plant Biol 13 : 36-41
Zhang Y, Giboulot A, Zivy M, Valot B, Jamet E, Albenne C (2011) Combining various strategies to increase the coverage of the plant cell wall glycoproteome. Phytochemistry 72 : 1109-1123 (read)
2010
Guirimand G, Courdavault V, Lanoue A, Mahroug S, Guihur A, Blanc N, Giglioli-Guivarc’h N, St-Pierre B, Burlat V (2010) Strictosidine activation in Apocynaceae : towards a “nuclear time bomb” ? BMC Plant Biol 10 : 20
Guirimand G, Courdavault V, St-Pierre B, Burlat V (2010) Biosynthesis and regulation of alkaloids. In E Pua, M Davey, eds, Plant developmental biology – Biotechnological perspectives, vol 2. Springer-Verlag, Berlin Heidelberg, pp 139-160
Jamet E, Pont-Lezica R (2010) The puzzle of protein location in plant proteomics. In G Rancourt, ed, Proteomics : Methods, Applications and Limitations. Nova Science Publishers, Inc., Hauppauge, NY, USA. (read)
Lanoue A, Burlat V, Henkes GJ, Koch I, Schurr U, Rose USR (2010) De novo biosynthesis of defense root exudates in response to Fusarium attack in barley. New Phytol 185 : 577-588
Lanoue A, Burlat V, Schurr U, Rose US (2010) Induced root-secreted phenolic compounds as a belowground plant defense. Plant Signal Behav 5 : 1037-1038
Pont-Lezica R, Minic Z, San Clemente H, Roujol D, Jamet E (2010) Plant cell wall functional genomics : Novelties from proteomics. In M Osborne, ed, Advances in Genetics Research, vol 1. Nova Science Publishers, Inc, Hauppauge, NY, USA, pp 265-281. (read)
2009
Albenne C, Canut H, Boudart G, Zhang Y, San Clemente H, Pont-Lezica R, Jamet E (2009) Plant cell wall proteomics : mass spectrometry data, a trove for research on protein structure/function relationships. Mol Plant 2 : 977-989. (read)
Courdavault V, Burlat V, St-Pierre B, Giglioli-Guivarc’h N (2009) Proteins prenylated by type I protein geranylgeranyltransferase act positively on the jasmonate signalling pathway triggering the biosynthesis of monoterpene indole alkaloids in Catharanthus roseus. Plant Cell Rep 28 : 83-93
Guirimand G, Burlat V, Oudin A, Lanoue A, St-Pierre B, Courdavault V (2009) Optimization of the transient transformation of Catharanthus roseus cells by particle bombardment and its application to the subcellular localization of hydroxymethylbutenyl 4-diphosphate synthase and geraniol 10-hydroxylase. Plant Cell Rep 28 : 1215-1234
Jamet E, Roujol D, San Clemente H, Irshad M, Soubigou-Taconnat L, Renou J, Pont-Lezica R (2009) Cell wall biogenesis of Arabidopsis thaliana elongating cells : transcriptomics complements proteomics. BMC Genomics 10 : 505 (read)
Minic Z, Jamet E, San Clemente H, Pelletier S, Renou J, Rihouey C, Okinyo D, Proux C, Lerouge P, Jouanin L (2009) Transcriptomic analysis of Arabidopsis developing stems : a close-up on cell wall genes. BMC Plant Biol 9 : 17 (read)
Pont-Lezica RF (2009) Localizing proteins by tissue printing. Methods Mol Biol 536 : 75-88 (read)
San Clemente H, Pont-Lezica R, Jamet E (2009) Bioinformatics as a tool for assessing the quality of sub-cellular proteomic strategies and inferring functions of proteins : plant cell wall proteomics as a test case. Bioinform Biol Insights 3 : 15-28 (read)
2008
Irshad M, Canut H, Borderies G, Pont-Lezica R, Jamet E (2008) A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana : confirmed actors and newcomers. BMC Plant Biol 8 : 94 (lire)
Jamet E, Albenne C, Boudart G, Irshad M, Canut H, Pont-Lezica R (2008) Recent advances in plant cell wall proteomics. Proteomics 8 : 893-908 (read)
Jamet E, Boudart G, Borderies G, Charmont S, Lafitte C, Rossignol M, Canut H, Pont-Lezica R (2008) Isolation of plant cell wall proteins. Methods Mol Biol 425 : 187-201 (read)
Jamet E, Canut H, Albenne C, Boudart G, Pont-Lezica R (2008) Cell wall. In G Agrawal, R Rakwal, eds, Plant Proteomics : Technologies, Strategies, and Applications. Wiley, USA, pp 293-307. (read)
Jouili H, Bouazizi H, Rossignol M, Borderies G, Jamet E, El Ferjani E (2008) Partial purification and characterization of a copper-induced anionic peroxidase of sunflower roots. Plant Physiol Biochem 46 : 760-767 (read)
2007
Boudart G, Minic Z, Albenne C, Canut H, Jamet E, Pont-Lezica R (2007) Cell wall proteome. In S Samaj, J Thelen, eds, Plant proteomics. Springer, Berlin, pp 169-185 (read)
Minic Z, Jamet E, Négroni L, der Garabedian AP, Zivy M, Jouanin L (2007) A sub-proteome of Arabidopsis thaliana mature stems trapped on Concanavalin A is enriched in cell wall glycoside hydrolases. J Exp Bot 58 : 2503-2512 (read)
2006
Feiz L, Irshad M, Pont-Lezica R, Canut H, Jamet E (2006) Evaluation of cell wall preparations for proteomics : a new procedure for purifying cell walls from Arabidopsis hypocotyls. Plant Methods 2 : 10 (read)
Gouget A, Senchou V, Govers F, Sanson A, Barre A, Rougé P, Pont-Lezica R, Canut H (2006) Lectin receptor kinases participate in protein-protein interactions to mediate plasma membrane-cell wall adhesions in Arabidopsis. Plant Physiol 140 : 81-90 (read)
Jamet E, Canut H, Boudart G, Pont-Lezica R (2006) Cell wall proteins : a new insight through proteomics. Trends Plant Sci 11 : 33-39 (read)
2005
Aubourg S, Brunaud V, Bruyere C, Cock M, Cooke R, Cottet A, Couloux A, Dehais P, Deleage G, Duclert A, Echeverria M, Eschbach A, Falconet D, Filippi G, Gaspin C, Geourjon C, Grienenberger J-M, Houlné G, Jamet E, Lechauve F, Leleu O, Leroy P, Mache R, Meyer C, Nedjari H, Negrutiu I, Orsini V, Peyretaillade E, Pommier C, Raes J, Risler J-L, Rivière S, Rombault S, Rouzé P, Schneider M, Schwob P, Small I, Soumayet-Kampetanga G, Stankovski D, Toffano C, Tognolli M, Caboche M, Lecharny A (2005) GENEFARM, structural and functional annotation of Arabidopsis gene and protein families by a network of experts. Nucleic Acids Res 33 : D641-D646.
Boudart G, Jamet E, Rossignol M, Lafitte C, Borderies G, Jauneau A, Esquerré-Tugayé M, Pont-Lezica R (2005) Cell wall proteins in apoplastic fluids of Arabidopsis thaliana rosettes : identification by mass spectrometry and bioinformatics. Proteomics 5 : 212-221. (read)
Charmont S, Jamet E, Pont-Lezica R, Canut H (2005) Proteomic analysis of secreted proteins from Arabidopsis thaliana seedlings : improved recovery following removal of phenolic compounds. Phytochemistry 66 : 453-461. (read)
Masclaux F, Galaud J, Pont-Lezica R (2005) The riddle of the plant vacuolar sorting receptors. Protoplasma 226 : 103-108. (reas)
Masclaux F, Pont-Lezica R, Galaud J (2005) Relationship between allelic state of T-DNA and DNA methylation of chromosomal integration region in transformed Arabidopsis thaliana plants. Plant Mol Biol 58 : 295-303. (read)
Schantz M, Jamet E, Guitton A, Schantz R, Houlné G (2005) Functional analysis of the bell pepper knolle gene (cakn) promoter region in tobacco plants and in synchronized BY2 cells. Plant Sci 169 : 155-163.
2004
Guzzardi P, Genot G, Jamet E (2004) The Nicotiana sylvestris extensin gene, Ext 1.2A, is expressed in the root transition zone and upon wounding. Biochim Biophys Acta 1680 : 83-92.
Jamet E (2004) Bioinformatics as a critical prerequisite to transcriptome and proteome studies. J Exp Bot 55 : 1977-1979. (read)
Masclaux F, Charpenteau M, Takahashi T, Pont-Lezica R, Galaud J (2004) Gene silencing using a heat-inducible RNAi system in Arabidopsis. Biochem Biophys Res Commun 321 : 364-369.
Senchou V, Weide R, Carrasco A, Bouyssou H, Pont-Lezica R, Govers F, Canut H (2004) High affinity recognition of a Phytophthora protein by Arabidopsis via an RGD motif. Cell Mol Life Sci 61 : 502-509. (read)
2003
Borderies G, Jamet E, Lafitte C, Rossignol M, Jauneau A, Boudart G, Monsarrat B, Esquerré-Tugayé M, Boudet A, Pont-Lezica R (2003) Proteomics of loosely bound cell wall proteins of Arabidopsis thaliana cell suspension cultures : a critical analysis. Electrophoresis 24 : 3421-3432. (lire)
Laval V, Masclaux F, Serin A, Carrière M, Roldan C, Devic M, Pont-Lezica R, Galaud J (2003) Seed germination is blocked in Arabidopsis putative vacuolar sorting receptor (atbp80) antisense transformants. J Exp Bot 54 : 213-221
Salva I, Jamet E (2003) The tobacco Ext 1.4 extensin gene family is not regulated in cells proliferating under hormone control Plant Physiol Biochem 41 : 363-367 (read)
Theses
Adélaïde Jacq (2013-2016) University Paul Sabatier, Toulouse 3. Caractérisation fonctionnelle d’AtLTP2, une protéine de transfert de lipides impliquée dans le contrôle de l’intégrité de la cuticule chez Arabidopsis thaliana. (read)
Edith Francoz (2012-2015) University Paul Sabatier, Toulouse 3. Rôle de la famille multigénique des peroxydases pariétales de classe III dans le développement et la dynamique des parois cellulaires végétales. (read)
PhD thesis prize Henri Gaussen of the Académie des Sciences Inscriptions et Belles lettres of Toulouse (2016) (read)
Huan Nguyen-Kim (2011-2015) University Paul Sabatier, Toulouse 3. Recherche de la fonction de protéines riches en hydroxyproline dans les parois végétales. (read)
May Hijazi (2011) University Paul Sabatier, Toulouse 3. Caractérisation structurale et fonctionnelle d’AGP31, une glycoprotéine atypique de la paroi chez Arabidopsis thaliana. (read)
Muhammad Irshad (2008) University Paul Sabatier, Toulouse 3. Dynamique des protéines pariétales au cours de l’élongation cellulaire dans des hypocotyles étiolés d’Arabidopsis thaliana : approches protéomique et transcriptomique. (read)
Anne Gouget (2006) University Paul Sabatier, Toulouse 3. Etude fonctionnelle d’un récepteur lectine kinase (LecRK79), potentiel partenaire dans les contacts paroi-plasmalemme chez Arabidopsis thaliana.
lrsv.gestion@univ-tlse3.fr.
Phone
Standard : +33 5.34.32.38.01
Fax : +33 5.34.32.38.02
Find us
24, chemin de Borde-Rouge.BP 42617 Auzeville.
31326, Castanet-Tolosan. FRANCE