Our team
Bernard DUMAS
DR CNRS

Elodie GAULIN
Assistant Professor

Laurent CAMBORDE
Engineer CNRS

Maryam HASHEMI
PhD
maryam.hashemi@lrsv.ups-tlse.fr

Andreï KISELEV
PhD
andrei. kiselev@lrsv.ups-tlse.fr

Moustafa KHATIB
Post-doc
moustafa. khatib@lrsv.ups-tlse.fr

Marion HORTALA
Engineer CDD
Former members
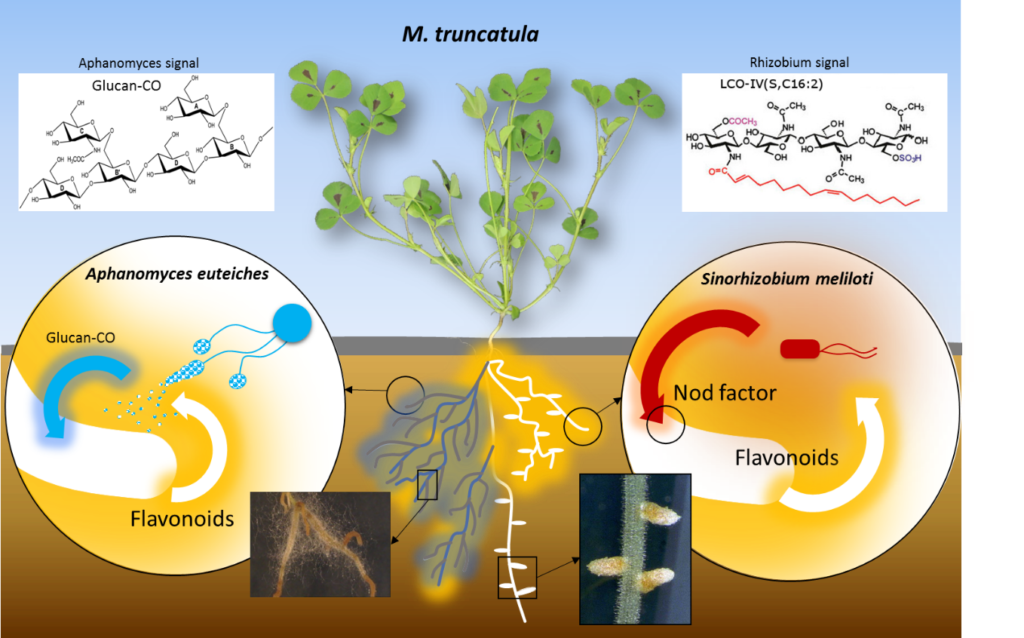
The model legume Medicago truncatula is well suited to study the interactions established in the root compartment with pathogenic or symbiotic microorganisms. Over the past years, the team has developed different approaches and methodologies based on the exploitation of genetic and genomic resources available for M. truncatula in order to:
1- study the genetic bases of quantitative resistance to Aphanomyces euteiches, the main parasite of pea in France and Europe.
2- analyse the connexions between immunity and symbiosis in legume species.
3- identify selection signatures in the genome of M. truncatula which could reflect evolutionary adaptation to root microorganisms.
4.. identify Aphanomyces genes playing a role in pathogenicity and adaptation to the host (effectors)
Plants, like animals, are exposed to a myriad of potential microbial pathogens, but diseases are still the exception, at least in natural ecosystems. Without any adaptive immune system, plants have developed an innate immune system that efficiently detects potentially dangerous microbes. One similarity between plant and animal innate immunity is that both are activated by the perception of “non-self” secreted Pathogen (or Microbial) Associated Molecular Patterns (PAMPs) through membrane-bound pattern recognition receptors (PRRs) at the plant cell surface. Responses to PAMPs include a set of biochemical and transcriptional responses leading to the first inducible layer of defense called PAMP-triggered immunity (PTI).
To establish infection, pathogens must evade, suppress, or otherwise manipulate PTI. This is achieved by the secretion of proteins called effectors which act either outside or inside plant cells to target and perturb signaling, regulatory or mechanistic processes associated with defense. Effectors may themselves be targeted by a second layer of defense called effector-triggered immunity (ETI), since their presence inside or outside host cells may be detected by resistance proteins (R genes). ETI is stronger than PTI and often accompanied by localized programmed cell death (hypersensitive response (HR)) to counteract the pathogen. Effectors that are recognized by the R proteins are termed avirulence (AVR) proteins.
The intracellular oomycete effectors
The completion of the genome sequence of several Phytophthora species including P. infestans and P. sojae has led to the identification of a huge and, rapidly diverging superfamily of proteins, encoded by 100-700 genes per genome. Proteins in this family use the N-terminal motif RXLR-EER to cross the host plasma membrane. Once inside the cells, the so-called RXLR proteins are presumed to modulate host defense signaling for the benefit of the parasite, and are thus referred to as effectors.
Another effector family called Crinklers (for Crinkling and Necrosis; CRN), has been also discovered in Phytophthora sp (> 200 genes). CRNs are modular proteins that comprise an N-terminal signal peptide, followed by a domain defined by the conserved but variable LXLFLAK motif involved in the translocation of CRN inside the host cell. The C-terminal domain is highly variable and is presumed to be involved in CRN function. So far, the function of CRN proteins is unknown.
The intracellular Aphanomyces euteiches CRN-effector family
Mining of the Aphanomyces genome revealed CRN genes, but not RXLR genes. A comparative approach is used to identify family genes associated with pathogenicity on particular hosts (plant, animals…). To this end, sequencing of several Aphanomyces has been completed. Genes coding for secreted proteins and induced during pathogenesis are functionally characterized.
2019
Vergnes S, Gayrard D, Veyssière M, Toulotte J, Martinez Y, DumontV, Bouchez O, Rey T, Dumas B (2019) Phyllosphere colonisation by a soil Streptomyces promotes plant defense responses against fungal infection Mol Plant Microbe Interact In press
Camborde L, Raynaud C, Dumas B, Gaulin E (2019) DNA-damaging effectors: new players in the effector arena. Trends Plant Sci. In press
Chervin J, Cabanac G, Audonnet M, Esquerré-Tugayé MT, Camborde L, Dumas B, Roux C, Talou T (2019) Deciphering the phylogeny of violets based on multiplexed genetic and metabolomic approaches Phytochemistry 163:99-110
Bonhomme M, Fariello MI, Navier H, Hajri A, Badis Y, Miteul H, Samac DA, Dumas B, Baranger A, Jacquet C, Pilet-Nayel ML. A local score approach improves GWAS resolution and detects minor QTL: application to Medicago truncatula quantitative disease resistance to multiple Aphanomyces euteiches Heredity 123: 517–531
Rey T, André O, Nars A, Dumas B, Gough C, Bottin A, Jacquet C. (2019) Lipo-chitooligosaccharide signalling blocks a rapid pathogen-induced ROS burst without impeding immunity. New Phytol. 221: 743-749
2018
Gaulin E, Pel JCM, Camborde L, San Clemente H, Courbier S, Dupouy MA, Lengelle J, Vayssiere M, Le Ru A, Grandjean F, Cordaux R, Moumen B, Gilbert C, Cano L, Aury JM, Guy J, Wincker P, Bouchez O, Klopp C, Dumas B (2018) Genomics analysis of Aphanomyces spp. identifies a new class of oomycete effector associated with host adaptation. BMC Biol. 16:43
Rey T, Jacquet C (2018) Symbiosis genes for immunity and vice versa. Curr. Opin. Plant Biol. 44:64-71
2017
Rey T, Bonhomme M, Chatterjee A, Gavrin A, Toulotte J, Yang W, André O, Jacquet C, Schornack S (2017) The Medicago truncatula GRAS protein RAD1 supports arbuscular mycorrhiza symbiosis and Phytophthora palmivora susceptibility. J. Exp. Bot. 68:5871-5881
Camborde L, Jauneau A, Brière C, Deslandes L, Dumas B, Gaulin E (2017) Detection of nucleic acid – protein interactions in plant leaves using fluorescence lifetime imaging microscopy. Nature Protocols 12 : 1933–1950
Rey T, Dumas B. (2017) Plenty is not plague : Streptomyces symbiosis with crops. Trends Plant Sci. 22:30-37
Gaulin E. (2017) Effector-mediated communication of filamentous plant pathogens with their hosts. Adv Bot Res 82:161-185
2016
Dufour MC, Magnin, N, Dumas B, Vergnes S, Corio-Costet MF (2016) High-throughput gene-expression quantification of grapevine defense responses in the field using Microfluidic Dynamic Arrays. BMC Genomics 17:957
Rey T, Laporte P, Bonhomme M, Jardinaud MF, Huguet S, Balzergue S, Dumas B, Niebel A, Jacquet C (2016) MtNF-YA1, a central transcriptional regulator of symbiotic nodule development, is also a determinant of Medicago truncatula susceptibility toward a root pathogen. Front Plant Sci 7 : 1837
Burgarella C, Chantret N, Gay L, Prosperi JM, Bonhomme M, Tiffin P, Young ND, Ronfort J. (2016) Adaptation to climate through flowering phenology : a case study in Medicago truncatula. Mol Ecol. 25:3397-415
Ramirez-Garces D, Camborde L, Pel MJ, Jauneau A, Martinez Y, Neant I, Leclerc C, Moreau M, Dumas B, Gaulin E. (2016) CRN13 candidate effectors from plant and animal eukaryotic pathogens are DNA-binding proteins which trigger host DNA damage response. New Phytol. 210:602-617.
2015
Martinez T, Texier H, Nahoum N, Lafitte C, Cioci G, Heux L, Dumas B, O’Donohue M, Gaulin E, Dumon C. (2015) Probing the functions of Carbohydrate Binding Modules in the CBEL protein from the Oomycete Phytophthora parasitica. PLoS One 10 : e0137481
Badis Y, Bonhomme M, Lafitte C, Huguet S, Balzergue S, Dumas B, Jacquet C (2015) Transcriptome analysis highlights preformed defenses and signaling pathways controlled by the prAe1 QTL, conferring partial resistance to Aphanomyces euteiches in Medicago truncatula. Mol. Plant.Pathol. 16 : 973-986
Bonhomme M, Boitard S, San Clemente H, Dumas B, Young N, Jacquet C (2015) Genomic signature of selective sweeps illuminates adaptation of Medicago truncatula to root-associated microorganisms. Mol. Biol. Evol. 2 : 2097–2110
Le Roux C, Huet G, Jauneau A, Camborde L, Tremousaygue D, Kraut A, Zhou B, Levaillant M, Adachi H, Yoshioka H, Raffaele S, Berthome R, Coute Y, Parker JE, Deslandes L (2015) A Receptor Pair with an Integrated Decoy Converts Pathogen Disabling of Transcription Factors to Immunity. Cell 161 : 1074-1088
Laffont C, Rey T, André O, Novero M, Kazmierczak T, Debellé F, Bonfante P, Jacquet C, Frugier F (2015) The CRE1 cytokinin pathway is differentially recruited depending on Medicago truncatula root environments and negatively regulates resistance to a pathogen. PLoS One. 10:e0116819
2014
Vergnes, S, Ladouce, N., Fournier, S., Ferhout, H., Attia, F., Dumas, B. (2014) Foliar treatments with Gaultheria procumbens essential oil induce defence responses and resistance against a fungal pathogen in Arabidopsis. Front. Plant Sci. 5:477
Bonhomme, M., André, O., Badis, Y., Ronfort, N., Burgarella, C., Chantret, N., Prosperi, J.M.,Briskine, R., Mudge, J., Debéllé, F., Navier, H., Miteul, H., Hajri, A., Baranger, A., Tiffin, P., Dumas, B., Pilet-Nayel, M.L., Young, N.D., Jacquet, C. (2014) High-density genome-wide association mapping identifies an F-box protein as the likely major component of Medicago truncatula resistance to Aphanomyces euteiches. New Phytol. 201:1328-42
2013
Nars A, Lafitte C, Chabaud M, Drouillard S, Mélida H, Danoun S, Le Costaouëc T, Rey T, Benedetti J, Bulone V, Barker DG, Bono JJ, Dumas B, Jacquet C, Heux L, Fliegmann J, Bottin A. (2013) Aphanomyces euteiches cell wall fractions containing novel glucan-chitosaccharides induce defense genes and nuclear calcium oscillations in the plant host Medicago truncatula. PLoS One 8:e75039
Larroque M, Belmas E, Martinez, M, Vergnes S, Ladouce N, Lafitte C, Gaulin E, Dumas B (2013) Pathogen-associated molecular pattern-triggered immunity and resistance to the root pathogen Phytophthora parasitica in Arabidopsis . J. Exp. Bot. 64 : 3615–3625
Gough C, Jacquet C (2013) Nod factor perception protein carries weight in biotic interactions. Trends Plant Sci. 18:566-574
Jiang RHY, de Bruijn I, Haas BJ, Belmonte R, Löbach L, Christie J, van den Ackerveken G, Bottin A, Bulone V, Dıaz-Moreno SM, Dumas B, Fan L, Gaulin E, Govers F, Grenville-Briggs LJ, Horner NR, Levin JZ, Mammella M, Meijer HJG, Morris P, Nusbaum C, Oome S, Phillips AJ, Rzeszutek E, van Rooyen D, Saraiva M, Secombes CJ, Seidl MF, Snel B, Stassen JHM, Sykes S, Tripathy S, van den Berg H, Vega-Arreguin JC, Wawra S, Young SK, Zeng Q, Dieguez-Uribeondo J, Russ C, Tyler BM, van West P (2013) Distinctive expansion of potential virulence genes in the genome of the oomycete fish pathogen Saprolegnia parasitica PLoS Genet. 9 : e1003272
Rey T, Nars A, Bonhomme M, Bottin A, Huguet S, Balzergue S, Jardinaud MF, Bono JJ, Cullimore J, Dumas B, Gough C, Jacquet C (2013) NFP, a LysM protein controlling Nod factor perception, also intervenes in Medicago truncatula resistance to pathogens. New Phytol. 198 : 875-886
Nars A, Rey T, Lafitte C, Vergnes S, Amatya S, Jacquet C, Dumas B, Thibaudeau C, Heux L, Bottin A, Fliegmann J (2013) An experimental system to study responses of Medicago truncatula roots to chitin oligomers of high degree of polymerization and other microbial elicitors. Plant Cell Rep. 32 : 489-502
2012
Larroque M, Barriot R, Bottin A, Barre A, Rouge P, Dumas B, Gaulin E (2012) The unique architecture and function of cellulose-interacting proteins in oomycetes revealed by genomic and structural analyses. BMC Genomics 13
2011
Jaulneau V, Lafitte C, Corio-Costet MF, Stadnik MJ, Salamagne S, Briand X, Esquerré-Tugayé, MT, Dumas B (2011) An Ulva armoricana extract protects plants against three powdery mildew pathogens Eur. J. Plant Pathol. 131 :393–401
Djébali N, Mhadhbi H, Lafitte C, Dumas B, Esquerré-Tugayé MT, Aouani ME and Jacquet C (2011) Hydrogen peroxide scavenging mechanisms are components of Medicago truncatula partial resistance to Aphanomyces euteiches Eur. J. Plant Pathol. 131 : 559-571
Larroque M, Ramirez D, Lafitte C, Borderies G, Dumas B, Gaulin E (2011) Expression and purification of a biologically active Phytophthora parasitica cellulose binding elicitor lectin in Pichia pastoris. Protein Expr Purif. 80 : 217-223 ![]()
Baxter L, Tripathy S, Ishaque N, Boot N, Cabral A, Kemen E, Thines M, Ah-Fong A, Anderson R, Badejoko W, Bittner-Eddy P, Boore JL, Chibucos MC, Coates M, Dehal P, Delehaunty K, Dong S, Downton P, Dumas B, Fabro G, Fronick C, Fuerstenberg SI, Fulton L, Gaulin E, Govers F, Hughes L, Humphray S, Jiang RH, Judelson H, Kamoun S, Kyung K, Meijer H, Minx P, Morris P, Nelson J, Phuntumart V, Qutob D, Rehmany A, Rougon-Cardoso A, Ryden P, Torto-Alalibo T, Studholme D, Wang Y, Win J, Wood J, Clifton SW, Rogers J, Van den Ackerveken G, Jones JD, McDowell JM, Beynon J, Tyler BM (2010) Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science 330: 1549-1551
Jaulneau V, Cazaux C, Wong Sak Hoi J, Fournier S, Esquerré-Tugayé MT, Jacquet C, Dumas B (2010) Host and non-host resistance in Medicago–Colletotrichum interactions. Mol Plant-Microbe Interact 23: 1107–1117
Fammartino A, Verdaguer B, Fournier J, Tamietti G, Carbonne F, Esquerré-Tugayé MT, Cardinale F (2010) Coordinated transcriptional regulation of the divinyl ether biosynthetic genes in tobacco by signal molecules related to defense. Plant Physiol Biochem 48: 225-231
Gaulin E, Bottin A, Dumas B (2010) Sterol biosynthesis in oomycete pathogens. Plant Signal Behav 5: 258-260
Jaulneau V, Lafitte C, Jacquet C, Fournier S, Salamagne S, Briand X, Esquerré-Tugayé MT, Dumas B (2010) Ulvan, a sulfated polysaccharide from green algae, activates plant immunity through the jasmonic acid signaling pathway. J Biomed Biotech 10: 525291
Levesque CA, Brouwer H, Cano L, Hamilton JP, Holt C, Huitema E, Raffaele S, Robideau GP, Thines M, Win J, Zerillo MM, Beakes GW, Boore JL, Busam D, Dumas B, Ferriera S, Fuerstenberg SI, Gachon CM, Gaulin E, Govers F, Grenville-Briggs L, Horner N, Hostetler J, Jiang RH, Johnson J, Krajaejun T, Lin H, Meijer HJ, Moore B, Morris P, Phuntmart V, Puiu D, Shetty J, Stajich JE, Tripathy S, Wawra S, van West P, Whitty BR, Coutinho PM, Henrissat B, Martin F, Thomas PD, Tyler BM, De Vries RP, Kamoun S, Yandell M, Tisserat N, Buell CR (2010) Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol 11: R73
Schornack S, van Damme M, Bozkurt TO, Cano LM, Smoker M, Thines M, Gaulin E, Kamoun S, Huitema E (2010) Ancient class of translocated oomycete effectors targets the host nucleus. Proc Natl Acad Sci USA 107: 17421-17426
Wong Sak Hoi J, Dumas B (2010) The STE12 and STE12-like proteins: fungal transcription factors regulating development and pathogenicity. Eukaryot Cell 9: 480-485
2009
Djebali N, Jauneau A, Ameline-Torregrosa C, Chardon F, Jaulneau V, Mathe C, Bottin A, Cazaux M, Pilet-Nayel ML, Baranger A, Aouani ME, Esquerré-Tugayé MT, Dumas B, Huguet T, Jacquet C (2009) Partial resistance of Medicago truncatula to Aphanomyces euteiches is associated with protection of the root stele and is controlled by a major QTL rich in proteasome-related genes. Mol Plant Microbe Interact 22: 1043-1055
Madoui MA, Bertrand-Michel J, Gaulin E, Dumas B (2009) Sterol metabolism in the oomycete Aphanomyces euteiches, a legume root pathogen. New Phytol 183: 291-300
Pilet-Nayel ML, Prosperi JM, Hamon C, Lesne A, Lecointe R, Le Goff I, Herve M, Deniot G, Delalande M, Huguet T, Jacquet C, Baranger A (2009) AER1, a major gene conferring resistance to Aphanomyces euteiches in Medicago truncatula. Phytopathology 99: 203-208
Tollot M, Wong Sak Hoi J, van Tuinen D, Arnould C, Chatagnier O, Dumas B, Gianinazzi-Pearson V, Seddas PM (2009) An STE12 gene identified in the mycorrhizal fungus Glomus intraradices restores infectivity of a hemibiotrophic plant pathogen. New Phytol 181: 693-707
2008
Ameline-Torregrosa C, Cazaux M, Danesh D, Chardon F, Cannon SB, Esquerre-Tugaye MT, Dumas B, Young ND, Samac DA, Huguet T, Jacquet C (2008) Genetic dissection of resistance to anthracnose and powdery mildew in Medicago truncatula. Mol Plant Microbe Interact 21: 61-69
Badreddine I, Lafitte C, Heux L, Skandalis N, Spanou Z, Martinez Y, Esquerré-Tugayé MT, Bulone V, Dumas B, Bottin A (2008) Cell wall chitosaccharides are essential components and exposed patterns of the phytopathogenic oomycete Aphanomyces euteiches. Eukaryot Cell 7: 1980-1993
Dumas B, Bottin A, Gaulin E, Esquerre-Tugaye MT (2008) Cellulose-binding domains: cellulose associated-defensive sensing partners? Trends Plant Sci 13: 160-164
Gaulin E, Madoui MA, Bottin A, Jacquet C, Mathé C, Couloux A, Wincker P, Dumas B (2008) Transcriptome of Aphanomyces euteiches: new oomycete putative pathogenicity factors and metabolic pathways. PLoS ONE 3: e1723
2007
Fammartino A, Cardinale F, Gobel C, Mene-Saffrane L, Fournier J, Feussner I, Esquerre-Tugaye MT (2007) Characterization of a divinyl ether biosynthetic pathway specifically associated with pathogenesis in tobacco. Plant Physiol 143: 378-388
Gaulin E, Haget N, Khatib M, Herbert C, Rickauer M, Bottin A (2007) Transgenic sequences are frequently lost in Phytophthora parasitica transformants without reversion of the transgene-induced silenced state. Can J Microbiol 53: 152-157
Gaulin E, Jacquet C, Bottin A, Dumas B (2007) Root rot disease of legumes caused by Aphanomyces euteiches. Mol Plant Pathol 8: 539-548
Madoui MA, Gaulin E, Mathé C, San Clemente H, Couloux A, Wincker P, Dumas B (2007) AphanoDB: a genomic resource for Aphanomyces pathogens. BMC Genomics 8: 471
Djebali N, Mhadhbi H, Jacquet C, Huguet T, Aouani ME (2007) Involvement of hydrogen peroxide, peroxidase and superoxide dismutase in response of Medicago truncatula lines differing in susceptibility to Phoma medicaginis infection. J. Phytopathol 155: 633-640
Wong Sak Hoi JW, Herbert C, Bacha N, O’Connell R, Lafitte C, Borderies G, Rossignol M, Rougé P, Dumas B (2007) Regulation and role of a STE12-like transcription factor from the plant pathogen Colletotrichum lindemuthianum. Mol Microbiol 64: 68-82
2006
Ameline-Torregrosa C, Dumas B, Krajinski F, Esquerre-Tugaye MT, Jacquet C (2006) Transcriptomic approaches to unravel plant-pathogen interactions in legumes. Euphytica 147 : 25-36
Gaulin E, Dramé N, Lafitte C, Torto-Alalibo T, Martinez Y, Ameline-Torregrosa C, Khatib M, Mazarguil H, Villalba-Mateos F, Kamoun S, Mazars C, Dumas B, Bottin A, Esquerré-Tugayé MT, Rickauer M (2006) Cellulose binding domains of a Phytophthora cell wall protein are novel pathogen-associated molecular patterns. Plant Cell 18 : 1766-1777
2005
Boudart G, Jamet E, Rossignol M, Lafitte C, Borderies G, Jauneau A, Esquerré-Tugayé MT, Pont-Lezica R (2005) Cell wall proteins in apoplastic fluids of Arabidopsis thaliana rosettes : identification by mass spectrometry and bioinformatics. Proteomics 5 : 212-221
Colditz F, Braun HP, Jacquet C, Niehaus K, Krajinski F (2005) Proteomic profiling unravels insights into the molecular background underlying increased Aphanomyces euteiches-tolerance of Medicago truncatula. Plant Mol Biol 59 : 387-406
Prost I, Dhondt S, Rothe G, Vicente J, Rodriguez MJ, Kift N, Carbonne F, Griffiths G, Esquerre-Tugaye MT, Rosahl S, Castresana C, Hamberg M, Fournier J (2005) Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol 139 : 1902-1913
Siriputthaiwan P, Jauneau A, Herbert C, Garcin D, Dumas B (2005) Functional analysis of CLPT1, a Rab/GTPase required for protein secretion and pathogenesis in the plant fungal pathogen Colletotrichum lindemuthianum. J Cell Sci 118 : 323-329
2004
Cluzet S, Torregrosa C, Jacquet C, Lafitte C, Fournier J, Mercier L, Salamagne S, Briand X, Esquerré-Tugayé MT, Dumas B (2004) Gene expression profiling and protection of Medicago truncatula against a fungal infection in response to an elicitor from the green algae Ulva spp. Plant Cell Environ 27 : 917-928
Herbert C, O’Connell R, Gaulin E, Salesses V, Esquerre-Tugaye MT, Dumas B (2004) Production of a cell wall-associated endopolygalacturonase by Colletotrichum lindemuthianum and pectin degradation during bean infection. Fungal Genet Biol 41 : 140-147
Khatib M, Lafitte C, Esquerré-Tugayé MT, Bottin A, Rickauer M (2004) The CBEL elicitor of Phytophthora parasitica var. nicotianae activates defence in Arabidopsis thaliana via three different signalling pathways. New Phytol. 162 : 501-510
O’Connell R, Herbert C, Sreenivasaprasad S, Khatib M, Esquerré-Tugayé MT, Dumas B (2004) A novel Arabidopsis–Colletotrichum pathosystem for the molecular dissection of plant-fungal interactions. Mol Plant Microbe Interact 17 : 272-282
Torregrosa C, Cluzet S, Fournier J, Huguet T, Gamas P, Prosperi JM, Esquerré-Tugayé MT, Dumas B, Jacquet C (2004) Cytological, genetic, and molecular analysis to characterize compatible and incompatible interactions between Medicago truncatula and Colletotrichum trifolii. Mol Plant Microbe Interact 17 : 909-920
2003
Borderies G, Jamet E, Lafitte C, Rossignol M, Jauneau A, Boudart G, Monsarrat B, Esquerré-Tugayé MT, Boudet A, Pont-Lezica R (2003) Proteomics of loosely bound cell wall proteins of Arabidopsis thaliana cell suspension cultures : a critical analysis. Electrophoresis 24 : 3421-3432
Boudart G, Charpentier M, Lafitte C, Martinez Y, Jauneau A, Gaulin E, Esquerré-Tugayé MT, Dumas B (2003) Elicitor activity of a fungal endopolygalacturonase in tobacco requires a functional catalytic site and cell wall localization. Plant Physiol 131 : 93-101
Mène-Saffrané L, Esquerré-Tugayé MT, Fournier J (2003) Constitutive expression of an inducible lipoxygenase in transgenic tobacco decreases susceptibility to Phytophthora parasitica var. nicotianae. Mol Breeding 12 : 271-282
Ongoing PhDs
M. Hashemi Effect of mycoparasite and root endophyte Pythium oligandrum on plant root-microbe interactions
A. Kiselev Molecular mechanisms of the interaction of the oomycete Aphanomyces euteiches with its environment and its plant host
C. Nicolle Rôle des Streptomyces racinaires dans le fonctionnement du microbiote racinaire
Completed PhDs
L. Camborde (2020) Caractérisation fonctionnelle e différents effecteurs candidats de l’oomycète Aphanomyces euteiches, parasite racinaire des légumineuses
C. Faure (2020) Mécanismes cytologiques et moléculaires du pouvoir pathogène de Pythium oligandrum, oomycète mycoparasite
D. Gayrard (2020) Activités stimulatrices des défense de plantes et antifongique d’une souche de Streptomyces sp. utilisée en tant qu’agent de biocontrôle
T. Martinez (2015) Les oomycètes, microorganismes pathogènes de plantes : nouvelles ressources biotechnologiques pour l’exploitation de la biomasse ligno-cellulosique
Y. Badis, (2014) Déterminants génétiques de la résistance de Medicago truncatula à Aphanomyces euteiches
D. Ramirez, (2014) Analyse des effecteurs Crinklers (CRN) de l’oomycète Aphanomyces euteiches.
O. André (2014) Analyse génétique de la résistance quantitative de Medicago truncatula au parasite racinaire Aphanomyces euteiches.
A. Nars, (2013) Caractérisation structurale et perception par la plante hôte Medicago truncatula des chitosaccharides pariétaux d’Aphanomyces euteiches, parasite de légumineuses. Lire
T. Rey, (2013) Mise en évidence et caractérisation d’interconnexions moléculaires entre symbiose et résistance chez la légumineuse modèle Medicago truncatula
M. Larroque, (2008-2011) Mécanismes de surveillance de l’intégrité pariétale et rôle dans l’induction des défenses des plantes. Lire
V. Jaulneau, (2010) Mode d’action de nouvelles molécules stimulatrices des défenses des plantes. (Confidentielle)
M. Cazaux, (2009) Analyse génétique de la résistance de la légumineuse Medicago truncatula à Colletotrichum trifolii. Lire
M.A. Madoui, (2009) Génomique fonctionnelle de l’interaction Medicago truncatula/Aphanomyces euteiches Lire
I. Badreddine, (2008) Etudes des surfaces cellulaires d’Aphanomyces euteiches parasite de légumineuses.Lire
N. Djébali, (2008) Etude des mécanismes de résistance de la plante modèle Medicago truncatula vis-à-vis de deux agents pathogènes majeurs des légumineuses cultivées : Phoma medicaginis et Aphanomyces euteiches. Lire
J. Wong Sak Hoi, (2007). Analyse fonctionnelle du facteur de transcription CLSTE12, impliqué dans le pouvoir pathogène du champignon phytopathogène Colletotrichum lindemtuhianum.
P.Siriputthaiwan, (2005). Sécrétion de protéines et pouvoir pathogène des champignons parasites de plantes : rôle d’une Rab/GTPase impliquée dans le transport vésiculaire.
C. Torregrosa, (2004) Caractérisation cytologique, génétique et moléculaire de l’interaction Colletotrichum trifolii/Medicago truncatula.
M. Khatib (2004) Expression par transgenèse d’un éliciteur de Phytophthora dans le Tabac et Arabidopsis et étude du phénotype.
C. Herbert (2003) Analyse moléculaire de la régulation de l’expression des gènes codant les endopolygalacturonases de Colletotrichum lindemuthianum, agent de l’anthracnose du Haricot.
lrsv.gestion@univ-tlse3.fr.
Phone
Standard : +33 5.34.32.38.01
Fax : +33 5.34.32.38.02
Find us
24, chemin de Borde-Rouge.BP 42617 Auzeville.
31326, Castanet-Tolosan. FRANCE